Authors: Shu Yi, Xu Jiaxin, Yang Chen, Chen Yilian, Luo Qian, Gong Bo, Yang Zhenglin, Huang Guo
Abstract [View PDF] [Read Full Text]
Objective
To detect whether Toll-like receptor 4 (TLR4) polymorphisms contributed to primary open angle glaucoma (POAG) in a Chinese population.
Methods
A Chinese cohort, including 799 unrelated POAG patients and 799 unrelated controls, was enrolled in our case-control association study. The data was collected at Sichuan Provincial People’s Hospital from May 2014 to March 2018. TLR4 functional single nucleotide polymorphisms (SNPs), including rs4986790 and rs4986791, were genotyped by SNaPshot method. Genotype and allele frequencies of the two SNPs were evaluated. This study was approved by the Institutional Review Boards of the Sichuan Academy of Medical Sciences & Sichuan Provincial People’s Hospital (No.2016-58), and complied with the guidelines of the Declaration of Helsinki. Written informed consents were obtained from all subjects prior to the study.
Results
Allelic association analysis revealed that there were no significant association detected in the allelic distributions between the POAG cases and controls for SNPs rs4986790 (P=0.317) and rs4986791 (OR=1.000, 95%CI =0.062 5-16.002 2, P=1.000) in the TLR4 gene. Conditional analysis of the two SNPs did not show any significant difference in genotype and allele frequency between the case and the control groups. No association of the two SNPs with POAG was detected under four different genetic models, including homozygote, heterozygote, dominant and recessive models.
Conclusions
Polymorphisms rs4986790 and rs4986791 in the TLR4 gene are not related to POAG in the Chinese cohort.
Key words:
Contributor Information
Glaucoma is characterized by progressive degeneration of retinal ganglion cell (RGC), followed by visual field defects and optic disc damage[1]. The estimated number with primary glaucoma is expected to rise to 76 million in 2020 and 111.8 million in 2040[2]. According to the etiology, glaucoma can be mainly divided into two broad categories: angle-closure glaucoma and open angle glaucoma. Primary open angle glaucoma (POAG) is a major type of glaucoma, accounting for about 68.6% of all glaucoma cases, and the incidence rate is increasing with age[1,3]. The progress of POAG is usually painless and patients may experience severe irreversible vision loss at an early stage, so it is very critical to determine new strategies for early diagnosis or even predict its risk.
The pathogenesis of POAG has been known to be multifactorial and complex. Common risk factors include ethnicity, advanced age, high myopia, diabetes, hypertension, smoking history and family history[4,5,6]. So far, genome-wide association analysis (GWAS) has identified that at least 20 genes or chromosomal loci are associated with POAG, such as TMCO1, CAV1-CAV2, ABCA1, CDKN2B-AS1, ARHGEF12, and SIX1-SIX6[7,8,9,10,11]. Recently, glaucoma has been recognized as an autoimmune disease, suggesting that autoimmunity may also be one of the causative factors of glaucoma[12]. Autoimmunity is considered to be the key to trabecular meshwork (TM) cell fibrosis and RGC death[13,14,15,16,17]. Several studies revealed that Toll-like receptor 4 (TLR4), expressed on the TM and RGC of POAG, is a common role in the innate immunity and contributes to the pathogenesis of glaucoma[9,18,19,20,21,22].
TLRs are a comparatively conservative family of receptors, which can specifically discern pathogen-associated molecular patterns[23]. TLRs have been found at least 13 members in mammals, and 11 of them are present in humans[24]. TLR4 has been the most investigated in the TLR family, which is a transmembrane receptor that can interact with exogenous ligands during inflammatory responses. In addition, TLR4 is not only implicated in bacterial lipopolysaccharides (LPSs) recognition, but also in endogenous ligands such as heat shock proteins (HSPs)[25,26]. In 2008, Shibuya et al. found that multiple single nucleotide polymorphisms (SNPs) in TLR4 gene were related to the risk of normal tension glaucoma (NTG) in the Japanese population[27]. Another association analysis was followed by Takano et al. also found evidence of association between TLR4 polymorphisms and NTG in 2012, suggesting thatTLR4 may play an important role in the pathogenesis of POAG[28].
A recent study manifested that two SNPs of the TLR4 gene, rs4986790 and rs4986791, carried an increased risk for POAG in Mexican population[29]. However, this result is different from the findings observed in a small sample of Saudi populations which could not detect the association between SNPs rs4986790 and rs4986791 of TLR4 and POAG[30]. In this study, we aimed to explore the possible association between rs4986790 and rs4986791 of theTLR4 gene and POAG in a Chinese Han population.
1 Materials and methods
1.1 Participants
This study was approved by the Institutional Review Boards of the Sichuan Academy of Medical Sciences & Sichuan Provincial People’s Hospital (No.2016-58) and complied with the guidelines of the Declaration of Helsinki. Written informed consents were obtained from all subjects prior to the study. The data was collected at Sichuan Provincial People’s Hospital from May 2014 to March 2018. The subjects in this research are all Han Chinese who lived in the Sichuan Province in Mainland China, and relatives of more than three generations are also Han Chinese. Subjects from other ethnic groups are excluded from this study. Sichuan Academy of Medical Sciences & Sichuan Provincial People’s Hospital recruited the samples of unrelated POAG and control subjects.
POAG diagnostic criteria were applied including the absence of any secondary form of glaucoma, such as exfoliation syndrome or a history of ocular trauma;gonioscopically open anterior chamber angle (Shaffer Grade Ⅲ or Grade Ⅳ);optic nerve damage, vertical cup-to-disc ratio (VCDR) higher than 0.6 or asymmetry in disc appearance, optic disc hemorrhage, and focal loss of the nerve fiber layer;characteristic visual field changes, and intraocular pressure (IOP) greater than 22 mmHg (1 mmHg=0.133 kPa) in both eyes without medications. Goldmann applanation tonometry (Haag Streit, Bern, Switzerland) was used to measure intraocular pressure (IOP) of all the included subjetcts, and visual field was tested by standard automated perimetry (Humphrey Field Analyzer Ⅱ, Carl Zeiss Meditec, Dublin, CA)[1]. In the study, unrelated control subjects were recruited from participants who attended the eye clinic for conditions, such as senile cataract, floaters, and mild refractive errors, were also excluded from this study, in order to be more rigorous. Controls underwent complete ophthalmic examinations. The inclusion criteria for controls were: (1)Chinese Han ancestry;(2)age at enrollment of more than 40 years;(3)IOP of less than 22 mmHg;(4)VCDR greater than 0.6;(5)an axial length of between 21.0 and 27.0 mm in both eyes;(6)absence of a family history of glaucoma;(7)absence of a history of glaucoma or elevated IOP;(8)absence of NTG;(9)absence of eye diseases that may severely affect visual acuity or visual field, such as retinitis pigmentosa, proliferative diabetic retinopathy, age-related macular degeneration, choroidal neovascularization, and pathological myopia. Compared to the control group, the mean age was younger, the mean IOP was higher, and the cup to disc ratio was larger in the case group, with significant differences between them (all at P<0.01)(Table 1).

Clinical features between the case group and control group (mean±SD)
1.2 DNA and genotyping
Venous blood from all the subjects was drawn and collected in an EDTA tube. The blood samples were preserved at 80 ℃ before genomic DNA extraction. Genomic DNA was extracted from the blood by serial phenol/chloroform extraction and ethanol precipitation. SNP genotyping was performed with the dye terminator-based SNaPshot method (Applied Biosystems, Foster City, CA). SNP analysis was performed on the ABI 3130 Genetic Analyzer (Applied Biosystems). Primers for polymerase chain reactions and SNaPshot were shown in Table 2. In brief, the polymerase chain reactions (10.0 μl final volume) contained 50 ng of genomic DNA, 1.0 μl of each primer (10 pmol/μl), 1.0 μl of 10× buffer (Takara Bio Inc., Shiga, Japan), 0.8 μl of deoxyribonucleotide triphosphates (2 mmol/L;Takara Bio Inc.), 0.4 μl MgCl2 (2.5 mmol/L;Takara Bio Inc.), and 0.1 μl of ExTaq polymerase (5 U/μl;Takara Bio Inc.). The product was then processed according to the ABI SNaPshot protocol.

Primers used for genotyping the two SNPs in the TLR4 gene
1.3 Statistical analysis
The Hardy-Weinberg equilibrium (HWE) for each SNP polymorphism, allelic or genotypic frequencies between the cases and controls were tested by the χ2 test using the software SPSS 23.0 (SPSS Inc., Chicago, IL). In terms of inferential analysis, odds ratios (OR), 95% confidence intervals (CI) and the corresponding P values were used to evaluate the association between different genotypes and the presence of disease. For the significance threshold of Pvalues, Bonferroni correction was applied to adjust the P values by the number of strata.P value <0.05 was considered statistically significant.
2 Results
SNPs rs4986790 (A/G) and rs4986791 (C/T) in the TLR4 gene were genotyped in all subjects. Since no genotype GG of rs4986790 was detected in POAG cases, HWE could not be calculated, and the allelic distribution of rs4986791 was in HWE (P>0.05). This result showed that there was no obvious genotyping error found in the genotyping method of this study, the genotypes of the functional SNPs fit the general population distribution, and the samples were randomly selected within a geographic region. The G allele frequency for rs4986790 was 0.001 in POAG cases and 0 in the control (P=0.317), respectively. The T allele frequency for rs4986791 was 0.001 in the POAG cases and the control (OR=1.000, 95% CI=0.062 5-16.002 2, P=1.000), respectively. Conditional analysis of the two SNPs showed no significant difference in genotype and allele frequency between the case and control groups, suggesting that there was no significant association between POAG and TLR4. In addition, we also tested the association of these two SNPs by using four different genetic models (homozygote, heterozygote, dominant and recessive models). However, rs4986790 and rs4986791 did not show any association with POAG in all four genetic models (Table 3, Table 4).
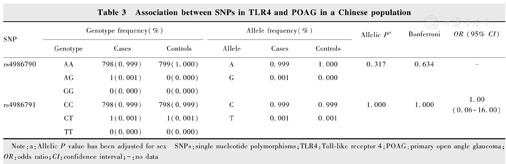
Association between SNPs in TLR4 and POAG in a Chinese population

Association analysis using four genetic models
3 Discussion
TLR is an important player in innate immune system and is deemed to be the bridge between innate immunity and adaptive immunity, which is associated with many autoimmune and allergic diseases, involving rheumatoid arthritis, bronchial asthma, septic shock, and allograft rejection[31,32,33,34,35]. TLR4 is the first TLR to be discovered which mainly identifying LPS on the cell wall of Gram-negative bacteria. By binding with LPS, TLR4 activates two main signaling pathways including myeloid differentiation factor 88 (MyD88) dependency and non-dependency. The former is mediated by MyD88 and adaptor proteins containing the TIR domain. MyD88-dependent pathway results in the activation of nuclear factor-kappaB (NF-κB) and the expression of pro-inflammatory genes[36]. The latter is mediated by TIR-containing proteins and TRIF. The initiation of the TRIF-dependent pathway leads to the activation of interferon regulatory factor and promotes the expression of interferon-β and interferon-inducible genes[37].
The human TLR4 gene is located on chromosome 9q33.1 and consists of 3 exons[38]. The function and activity of the product are mainly affected by genetic variation. SNPs s4986790 and rs4986791 in the TLR4 gene affect the extracellular domain of the protein. The genetic variation of the missense in the TLR4 polypeptide caused by the coding mutation is related to the low reactivity of the receptor[39]. Interestingly, compared to the rs4986790/rs4986791 haplotype individual, the rs4986790 haplotype showed a strong pro-inflammatory TNF-cytokine response after stimulation with LPS[40].
We analyzed the involvement of two reported functional TLR4 polymorphisms of POAG in a Chinese population in this study. A case-control study of two TLR4 functional SNPs showed the lack of significant association between rs4986790 and rs4986791 and POAG. The association of rs4986790 had the same trend of that in the Saudi Arabian population, but was different from Mexican population.
According to our statistics, the distribution of SNP rs4986790 of the TLR4 gene is population-specific. The mutation rate of rs4986790 in African population is the highest with an average of about 16% and the second is Caucasian white. In Asia, The incidence of mutation of rs4986790 is low in Chinese Han population[41,42]. In addition, in another study by Chen et al., TLR4 SNP rs7037117 was reported to be associated with late-onset POAG in southern China, but not in northern China[43]. Therefore, the association of TLR4 polymorphisms with POAG may require further evaluation in a larger cohort.
However, there is no direct evidence to support the activation, expression and signal transduction of TLR4 are implicated in glaucoma at present, but different findings have revealed its relationship to glaucoma pathology. In human glaucoma donor eyes, elevated IOP can cause up-regulation of TLR2, TLR3 and TLR4 and neurodegenerative enhancement by upregulation of TLR4 and complement system pathways has been confirmed in animal models[44,45,46]. Non-coding regions of the TLR4 polymorphism, including 50 untranslated regions, 30 untranslated regions and introns have also been shown to be related to POAG in Japanese population[28,29].
In summary, we did not find any direct link between the genotype or allele frequency of SNP rs4986790 and rs4986791 of the TLR4 gene and POAG in the Chinese Han cohort. Future studies may explain the missing link between the genetic mechanism of innate immunity and glaucoma in adults.
Conflicts of interest The authors have no conflicts of interest. The authors alone are responsible for the content and writing of the paper
References
.Expresstion of the Toll-like receptor 4 in rat retina with chronic ocular hypertention[J]. Chin J Exp Ophthalmol,2011,29(5):407–411.DOI:10.3760/cma.j.issn.2095-0160.2011.05.006.