·Clinical Research·
Evaluation of plasma exosomal miR-214-3p as a potential diagnostic and prognostic biomarker of uveal melanoma
Zhou Wenda, Shao Lei, Dong Li, Shi Xuhan, Zhang Ruiheng, Li Heyan, Wu Haotian, Wei Wenbin
Beijing Tongren Eye Center, Beijing Key Laboratory of Intraocular Tumor Diagnosis and Treatment, Beijing Ophtahlmology & Visual Sciences Key Laboratory, Medical Artificial Intelligence Research and Verification Laboratory of the Ministry of Industry and Information Technology, Beijing Tongren Hospital, Capital Medical University, Beijing 100730, China
Corresponding author: Wei Wenbin, Email: weiwenbintr@163.com
Abstract [View PDF in English] [View PDF in Chinese] [Read Full Text]
Objective To investigate the differential expression of miR-214-3p in plasma exosomes in different histological types of uveal melanomas (UMs) and evaluate miR-214-3p as a potential diagnostic and prognostic biomarker of UM.
Methods The study cohort included 25 UM patients (10 with spindle-shaped melanomas, 10 with epithelioid cell melanomas, and five with metastatic UMs, including four with epithelioid cell melanoma and one with metastatic spindle-shaped melanoma) who underwent enucleation at Beijing Tongren Hospital (Capital Medical University, Beijing, China) between December 2015 and October 2019 and 10 healthy donors as normal controls. Blood samples were collected from all the subjects for isolation of plasma exosomes. The morphology of exosomes was examined under an electron microscope. Exsomal marker proteins were identified by western blot analysis. Expression levels of miR-214-3p in plasma exosomes were detected by real-time fluorescence quantitative polymerase chain reaction. Differential expression of miR-214-3p among patients with different histological types of UM and healthy controls was compared. The area under the receiver operating characteristic curve (AUC) was calculated to assess the diagnostic and classification performance of exosomal miR-214-3p.
Results The isolated exosomes were hemispherical in shape with one concave side. The diameter of the exosomes was about 100 nm and the mean particle diameter was 82.0 ± 2.7 nm. The exosomes were positive for tumor susceptibility gene 101 and negative for calnexin. Relative expression levels of plasma exosomal miR-214-3p of the healthy controls, in situ UM patients, and metastatic UM patients were 0.86 (0.57, 1.49), 0.24 (0.10, 0.67), and 0.43 (0.23, 0.56), respectively. Relative expression of miR-214-3p in plasma exosomes was significantly lower in patients with in situ UM and metastatic UM than the healthy controls (Z=2.62, P<0.01; Z=2.08, P<0.05). Relative expression levels of exosomal miR-214-3p in patients with spindle cell melanoma-like UM and epithelioid cell-like UM were 0.11 (0.07, 0.64) and 0.46 (0.14, 0.91), respectively. There was no significant difference in plasma exosomal miR-214-3p levels among the different histopathological types of UM (all P>0.05). The AUC of plasma exosomal miR-214-3p for diagnosis of UM was 0.795.
Conclusions Plasma exosomal miR-214-3p levels were significantly reduced in both in situ UM and metastatic UM. Plasma exosomal miR-214-3p is a potential diagnostic biomarker for UM, but insufficient to distinguish among histological types of UM.
Key words Uveal melanoma; Circulating MicroRNA; Biomarkers; Exosomes; Early diagnosis; Prognostic evaluation
Fund program: National Natural Science Foundation of China (82141128); Capital Health Research and Development of Special Grants (2020-1-2052); Science & Technology Project of Beijing Municipal Science & Technology Commission (Z201100005520045, Z181100001818003); Beijing Outstanding Talent Project Youth Backbone Foundation (2018000021469G207); The Priming Scientific Research Foundation for Junior Researchers of Beijing Tongren Hospital, Capital Medical University (2018-YJJ-ZZL-045)
DOI: 10.3760/cma.j.cn115989-20220321-00110
Uveal melanoma (UM) is the most common intraocular malignancy in adults. Due to the lack of objective quantitative biomarkers, the clinical diagnosis of UM is generally based on indirect ophthalmoscopy combined with magnetic resonance imaging and B-scan ultrasonography 1, 2. Pathological examination after enucleation remains the gold standard for diagnosis of UM. Although highly accurate, the application of pathological examination is limited by enucleation, especially with the development of plaque brachytherapy. Hence, a noninvasive, convenient, and quantitative biomarker for the diagnosis of UM is urgently needed.
UM tends to metastasize even after treatment in more than 50% of patients. However, there are relatively few options for treatment of metastatic UM 3-5. Early treatment of high risk UM can reduce the risk of metastasis 6. MicroRNAs (miRNAs) are small, non-coding, single-stranded RNA molecules that post-transcriptionally regulate gene expression by inhibiting or inactivating target messenger RNAs. Expression patterns of miRNAs are frequently dysregulated in human tumors 7. The expression levels of various circulating miRNAs are measured as biomarkers in plasma, saliva, urine, and lacrimal fluid (tears) 8. Our group previously identified circulating miR-199a-3p as diagnostic and prognostic biomarker of both early and advanced UM 9. Previous studies have found that miR-21-5p acts as oncogene in cancers of the stomach, lung, and colon 10-12, and inhibits expression of P53 and promotes expression of LASP1, which stimulates the proliferation and metastasis of UM 13-14. In contrast to miR-21-5p, the role of miR-214-3p in the onset and progression of UM remains unclear.
Our group previously reported significant down-regulation of miR-214-3p in UM 15 and a similar pattern for plasma miR-214-3p, suggesting that miR-214-3p acted as anti-oncogene in UM. However, comparisons of plasma samples from UM patients with different histological types found that plasma miR-214-3p was down-regulated in epithelioid cell melanoma and especially spindle-shaped melanoma, thereby raising questions about the role of miR-214-3p in UM.
Exosomes are extracellular vesicles with diameters of 40–100 nm present in almost body fluids, including plasma 16-18. In this study, expression levels of miR-214-3p were measured in plasma exosomes of UM as a potential diagnostic and prognostic biomarker of UM.
1 Methods and Materials
1.1.1 Study approval and patient consent The protocol of this cross-sectional study was approved by the Ethics Committee of Capital Medical University (approval no. TRECKY2018-056). All patients and healthy donors were informed of the purpose of the study and submitted signed informed consent prior to inclusion in this study.
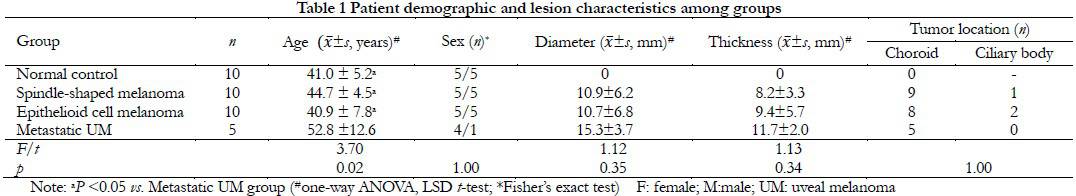
1.1.2 Baseline data Blood samples were collected from 25 UM patients (10 with spindle-shaped melanoma, 10 with epithelioid cell melanoma, and five with metastatic UM, including four with epithelioid cell melanoma and one with metastatic spindle-shaped melanoma) who underwent enucleation at Beijing Tongren Hospital (Capital Medical University, Beijing, China) between December 2015 and October 2019 and 10 healthy donors as normal controls. There were significant differences in age among the UM and healthy control groups (F=3.70, P<0.05) due to the advanced age of patients with metastatic UM. There was no significant difference in the ratio of males to females among the four groups (P=1.00) and no significant differences in tumor diameter and height among the UM groups (F=1.12, P=0.35, F=1.13, P=0.34). Most of the UM patients had choroid melanoma and there was no significant difference in tumor location among the three UM groups (P=1.00). Extraocular invasion was not observed in any of the patients (Table 1).
1.2 Methods
1.2.1 Plasma sample Fasting venous blood samples were collected in vacuum tubes from all patients and healthy controls. After 30 min at room temperature, the blood samples were centrifuged at 1,200 r/min for 15 min at 4°C. Then, plasma samples were collected and stored at -80℃. All patients underwent enucleation after blood collection. Histopathological examinations of the enucleated eyes were performed by the Department of Pathology of Beijing Tongren Hospital to determine the histopathological type in addition to the tumor location, diameter, height, and malignancy status.
1.2.2 Extraction of plasma exosomes Plasma exosomes were isolated by ultracentrifugation at 2,000 × g for 30 min at 4°C. Afterward, the supernatant was transferred to a new centrifuge tube and centrifuged at 10,000 × g for 45 min at 4°C to remove all larger vesicles. The supernatant was extracted and filtered through a membrane with a pore size of 0.45 μm. The filtrate was transferred to a new centrifuge tube and centrifuged at 100,000 × g for 70 min at 4°C. The supernatant was collected and centrifuged for 70 min. Finally, the exosomes were suspended in 10 mL of phosphate-buffered saline and stored at -80℃.
1.2.3 Exosome morphology Plasma exosome samples from each group were analyzed with a transmission electron microscope (HT-7700; Hitachi Ltd., Tokyo, Japan). In brief, each plasma exosome sample (10 μL) was applied to a copper mesh for 1 min and the floating liquid was absorbed by filter paper. Afterward, uranyl acetate (10 μL) was applied to the copper mesh for 1 min and the floating liquid was absorbed by filter paper. Each sample was dried at room temperature for several minutes and then imaged with a transmission electron microscope at 100 kV.
1.2.4 Analysis of particle size Two plasma exosome samples from each group were analyzed. In brief, 10 μL of each plasma exosome sample were diluted to 30 μL with phosphate-buffered saline and analysis of particle size was performed using a particle size analyzer (N30E; NanoFCM INC., Xiamen, China).
1.2.5 Identification of exosome biomarkers Two plasma exosome samples from each group were used for identification of proteins as biomarkers of exosomes. In brief, the exosomes samples were rapidly thawed at 37°C and lysed in a six-fold volume of radioimmunoprecipitation assay buffer on ice for 30 min. The protein concentration was determined with the bicinchoninic acid assay. After denaturation, the proteins were separated by electrophoresis and then transferred to a polyvinylidene fluoride membrane, which was blocked with 5% skim milk/Tris-buffered saline with 0.1% Tween® 20 detergent for 1 h and then probed with primary antibodies against tumor susceptibility gene 101 (ab125011; dilution, 1:1,000) and calnexin (ab22595; dilution, 1:1,000; Aibo Trading Co., Ltd., Shanghai, China) overnight at 4°C, followed by secondary antibodies (dilution, 1:5,000) at room temperature for 1 h. The protein bands were visualized with enhanced chemiluminescence reagent and imaged with a chemiluminescence imaging system (ChemiScope 3000mini; Clinx Science Instruments Co., Ltd., Shanghai, China).
1.2.6 RNA isolation Total RNA was extracted from each sample using the RNeasy Kit (Qiagen, Hilden Germany) in accordance with the manufacturer’s instructions. Each plasma exosome sample was transferred to an Eppendorf tube, spiked with 900 μL of lysis buffer, shaken vigorously for 30 s, and left at room temperature for 5 min to allow complete separation of the nucleoprotein complexes. Following the addition of 0.2 mL of chloroform, the tube was shaken vigorously for 15 s, left at room temperature for 5 min, then centrifuged at 12,000 r/min for 10 min. The aqueous phase was transferred to a new Eppendorf tube, mixed with a two-fold volume of absolute ethanol, added to an adsorption column, allowed to stand for 2 min, and then centrifuged at 12,000 r/min for 1 min. Afterward, 700 μL of deproteinized water were added to the adsorption column, which was allowed to stand for 2 min, then centrifuged at 12,000 rpm for 1 min. Then, 500 μL of rinse solution were added to the adsorption column, which was allowed to stand for 2 min, then centrifuged at 12,000 rpm for 1 min. After centrifugation at 12,000 rpm for 2 min, the collection tube was discarded, the adsorbent column was allowed to stand for 10 min, and the residual rinse solution in the adsorbent column was removed. Afterward, the adsorption column was transferred to a new tube and 15 μL of RNase-free ddH2O were added, then left at room temperature for 5 min and centrifuged at 12,000 rpm for 2 min to obtain the RNA sample.
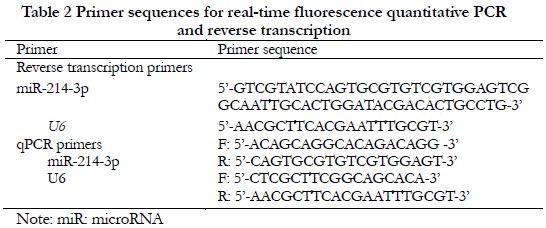
1.2.7 Quantification of miR-214-3p in plasma exosomes For quantification of miR-214-3p, template RNA was reverse-transcribed into complementary DNA (cDNA) using a universal reverse transcription kit (11141es60; Shanghai Yisheng Biotechnology Co., Ltd., Shanghai, China). The cDNA was amplified using a real-time polymerase chain reaction (PCR) fluorescent quantification kit (11201es08; Shanghai Yisheng Biotechnology Co., Ltd.) and a real-time PCR instrument (Ma-6000; Suzhou Yarui Biotechnology Co., Ltd., Suzhou, China). Each 20-μL PCR reaction consisted of 10 μL of PCR master mix, 1 μL of the appropriate primers, 5 μL of cDNA as a template, and 4 μL of sterile ultrapure water. The primer sequences are shown in Table 2. U6 was used as an internal reference. The reaction conditions included an initial denaturation step at 95°C for 5 min, followed by 40 cycles at 95°C for 10 s, 60°C for 20 s, and 72°C for 20 s. All reactions were run in triplicate. Relative expression levels were calculated with the 2-ΔΔCt method.
1.3 Statistical analysis
Statistical analyses were performed with IBM SPSS Statistics for Windows, version 25.0. (IBM Corporation, Armonk, NY, USA) (https://www.ibm.com/support/ pages/how-cite-ibm-spss-statistics-or-earlier-versions-spss). The Shapiro-Wilk test was used to confirm normal distribution of continuous variables. The data are presented as the mean ± standard deviation. Differences between groups were identified by one-way analysis of variance (ANOVA) followed by the least significant difference (LSD) test for pairwise comparisons between groups. Measured data with skewed distribution are presented as the median (interquartile range) and the significance of differences between groups was determined with the Kruskal–Wallis H test followed by the Mann-Whitney U test. Counted data are presented as the frequency and percentage and the significance of differences between groups was determined with the Fisher exact test. The specificity and sensitivity of circulating plasma exosomal mir-214-3p as diagnostic and prognostic biomarkers were assessed by calculating the area under the receiver operating characteristic curve (AUC). A probability (P) value of <0.05 was considered statistically significant.
2 Results
2.1 Identification of plasma exosomes
Images obtain with a transmission electron microscope showed that the exosomes were hemispherical with a concave side and an average diameter of about 100 nm. The mean particle size of the exosomes was 82.0 ± 2.7 nm. Western blot analysis confirmed that the exosomes were positive for tumor susceptibility gene 101 and negative for calnexin (Figure 1).
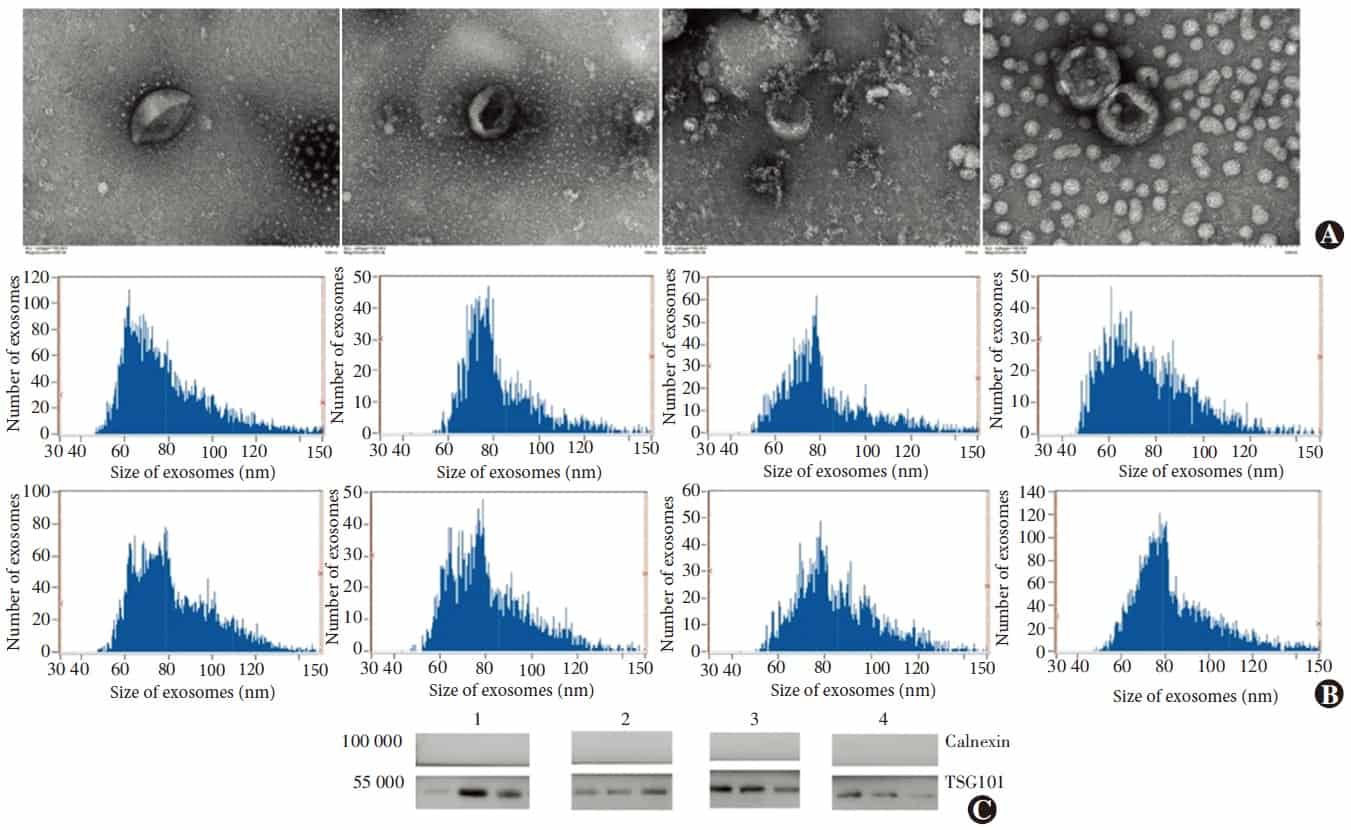
Figure 1 Identification of the exosomes A: Ultrastructure of the exosomes under transmission electron microscope The vesicle of the exosomes was integrated and hemispherical-like with a concavity inside B: Particle diameter analysis of the exosomes Vesicles showed a diameter from 79 to 84 nm, which was in the range of exosome size C: Expression of Exosomal marker protein TSG101 by Western blot Exosomal marker protein TSG101 was detected in all samples, and non-exosomal marker Calnexin was undetectable 1: Normal control; 2: Spindle-shaped melanoma; 3: Epithelioid cell melanoma; 4: Metastatic UM
2.2 Normalized levels of miR-214-3p in plasma exosomes
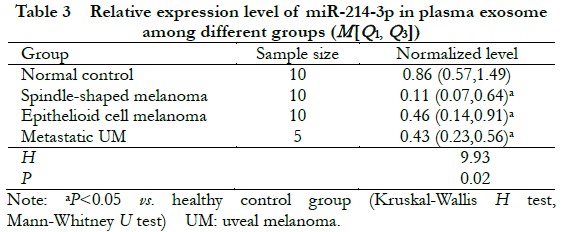
There were significant differences in normalized levels of microsomal miR-214-3p between the normal control group and all three UM groups (H=9.93, P=0.02). The normalized level of mir-214-3p was significantly higher in the normal control group than the three UM groups (all at P<0.05) (Table 3).
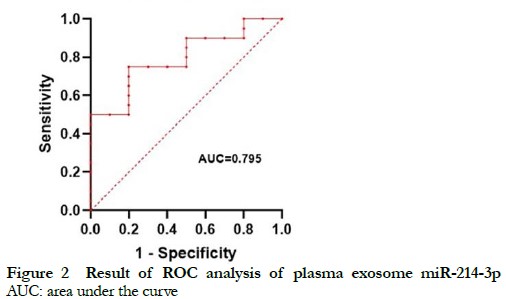
2.2.1 Performance of miR-214-3p in plasma exosomes as a diagnostic biomarker of UM The normalized level of miR-214-3p in plasma exosomes of the non-metastatic UM groups (including spindle-shaped melanoma and epithelioid cell melanoma) was 0.24 (0.10, 0.67), which was significantly lower than that of the normal control group of 0.86 (0.57, 1.49) (Z=2.62, P<0.01). The AUC as an indicator of the accuracy of miR-214-3p in plasma exosomes as a diagnostic biomarker of UM was 0.795 (Figure 2). Further comparisons of normalized levels of miR-214-3p in plasma
exosomes revealed significant differences between the normal control group and metastatic UM groups (Z=2.08, P<0.05).
2.2.2 Performance of miR-214-3p in plasma exosomes as a prognostic biomarker of UM There was no significant difference in the normalized levels of miR-214-3p in plasma exosomes between the spindle-shaped melanoma group and the epithelioid cell melanoma group (Z=1.66, P=0.09). Further comparisons of the normalized levels of miR-214-3p in plasma exosomes revealed no significant difference between the metastatic UM groups and epithelioid cell melanoma group (Z=0.84, P=0.80).
3 Discussion
MiRNAs, as non-coding RNAs, participate in the regulation of various cell functions by negatively regulating specific mRNAs and are related to the onset and progression of numerous diseases, including various cancers 19. In addition to the cytoplasm, miRNAs also exist in all body fluids, including the plasma 20. Circulating miRNAs mainly exists in three forms: bound to proteins in nucleic acid-protein complexes, bound to lipoproteins in lipoprotein-nucleic acid complexes, and in exosomes 21-23. Notably, miRNAs in exosomes are encapsulated and, thus, can stably exist in the blood and avoid decomposition by nucleases. Hence, miRNAs have received wide attention as potential non-invasive and quantifiable tumor biomarkers. In addition, exosomes actively packaged and secreted by tumor cells participate in intercellular signaling for regulation of the growth of tumor cells 18. The differential expression of miRNAs in exosomes is particularly significant to elucidate the molecular mechanisms involved in oncogenesis.
A previous study by our group found that miR-214-3p was significantly down-regulated in both spindle-shaped and epithelioid cell melanomas as compared to normal uveal tissues, while there were no significant difference between the two types of melanoma 15. However, miR-214-3p was significantly down-regulated in the plasma of UM patients. Notably, the AUC of plasma miR-214-3p as a diagnostic biomarker of UM was 0.775. The decrease in plasma miR-214-3p was more obvious in spindle-shaped melanoma than epithelioid cell melanoma. The AUC of plasma miR-214-3p to differentiate among histological types of UM was 0.930 9. The results of the present study revealed that the normalized level of miR-214-3p in plasma exosomes was significantly down-regulated in UM patients and the AUC of miR-214-3p in plasma exosomes as a diagnostic biomarker of UM was higher than that of plasma miR-214-3p, which is consistent with the results of our previous study. As compared to plasma, miRNAs in plasma exosomes are more resistant to nucleases. In addition, exosomes are actively secreted by cells, which can prevent the influence of miRNAs passively entering the blood during apoptosis. Therefore, miRNAs in plasma exosomes are more suitable for use as circulating biomarkers. The results of this study confirmed that the performance of miR-214-3p as a diagnostic biomarker of UM was superior to that of plasma miR-214-3p, demonstrating the potential of circulating miR-214-3p as a diagnostic biomarker of UM.
UM is classified into three distinct histological types: spindle-shaped melanoma, mixed melanoma, and epithelioid cell melanoma. The prognosis of spindle-shaped melanoma is generally better than that of epithelioid cell melanoma. Therefore, in this study, the usefulness of miR-214-3p in plasma exosomes as prognostic biomarker of UM was investigated.
The results of a previous study by our group showed that plasma miR-214-3p was significantly up-regulated in epithelioid cell melanoma as compared to spindle-shaped melanoma 9. Some miRNAs that function as tumor suppressors are selectively secreted by tumor cells, which promote the proliferation of tumor cells24. Previous studies have reported that miR-214-3p acts as a tumor suppressor in multiple cancers 25-26 and that epithelioid cell melanoma actively secretes greater amounts of miR-214-3p than spindle-shaped melanoma, thereby promoting proliferation of cancer cells. In this study, there was no statistically significant difference in normalized levels of miR-214-3p in plasma exosomes between spindle-shaped melanoma and epithelioid cell melanoma, which is consistent with the result of our previous study. Actually, blood cells are the main source of circulating miRNAs, although there are differences in levels of miRNAs in the blood versus tumor cells. However, it is more advantageous to monitor miRNAs in plasma exosomes, which are actively secreted by cells 24, 27. A particular miRNA can target hundreds of mRNAs, which explains why miR-214-3p is not only involved in tumor development, but other pathophysiological processes as well, including inflammation 28-29. Down-regulation of plasma miR-214-3p may be caused by other pathophysiological processes not necessarily associated with UM. In this study, normalized levels of miR-214-3p in plasma exosomes were significantly down-regulated in metastatic UM patients as compared to the normal controls, while there was no significant difference between metastatic UM and epithelioid cell melanoma, demonstrating that the circulating miR-214-3p may not be associated with UM. In addition, it should be emphasized that in addition to exosomes, circulating miRNAs also existed in the form of protein-nucleic acid complexes and lipoprotein-nucleic acid complexes, which are also actively secreted by cells 21-22. The results of this study did not completely negate the association between the circulating miR-214-3p and the prognosis of UM.
In this study, circulating miR-214-3p was identified as a potential biomarker for early diagnosis of UM. In addition, circulating miRNAs played important roles in intercellular communication and, thus, are useful to elucidate the mechanisms underlying tumor development.
Mir-214-3p can act as either a tumor suppressor gene or an oncogene 25-26, 30-32. The results of this study suggested that miR-214-3p acted as a suppressor gene in UM and reduced levels in blood and tumor tissue could be a mechanism related to the onset and progression of UM, suggesting that miR-214-3p is a potential target for treatment of UM.
As a potential limitation to this study, long-term follow-up of patients was not performed, thus survival analysis could not be conducted. In addition, circulating miR-214-3p in patients after treatment was not monitored and the patient cohort was relatively small. Therefore, future multicenter studies with larger cohorts across different races are warranted to further verify the association between circulating miR-214-3p and UM.
Conflict of interest None declared.
Author contributions Zhou Wenda: study concept, experimental design and implementation, data collection, analysis, and interpretation, and drafting of the article; Shao Lei and Li Dong: study concept, experimental design and implementation, data collection, analysis, and interpretation; Shi Xuhan, Zhang Ruiheng, Li Heyan, and Wu Haotian: experimental implementation, data collection, analysis, and interpretation; Wei Wenbin: study concept, experimental design, and final approval of the manuscript for publication.
References
[1] Fallico M, Raciti G, Longo A, et al. Current molecular and clinical insights into uveal melanoma (review)[J/OL]. Int J Oncol, 2021, 58(4):10[2022-03-01]. https://pubmed.ncbi.nlm.nih.gov/33649778/. DOI: 10.3892/ijo.2021.5190.
[2] Foti PV, Travali M, Farina R, et al. Diagnostic methods and therapeutic options of uveal melanoma with emphasis on MR imaging-Part I: MR imaging with pathologic correlation and technical considerations[J/OL]. Insights Imaging, 2021, 12(1):66[2022-03-01]. https://pubmed.ncbi.nlm.nih.gov/34080069/. DOI: 10.1186/s13244-021-01000-x.
[3] Kujala E, Mäkitie T, Kivelä T. Very long-term prognosis of patients with malignant uveal melanoma[J]. Invest Ophthalmol Vis Sci, 2003, 44(11):4651-4659. DOI: 10.1167/iovs.03-0538.
[4] Jensen OA. Malignant melanomas of the human uvea: 25-year follow-up of cases in Denmark, 1943–1952[J]. Acta Ophthalmol (Copenh), 1982, 60(2):161-82. DOI: 10.1111/j.1755-3768.1982.tb08371.x.
[5] Branisteanu DC, Bogdanici CM, Branisteanu DE, et al. Uveal melanoma diagnosis and current treatment options (review)[J/OL]. Exp Ther Med, 2021, 22(6):1428[2022-03-02]. https://pubmed.ncbi.nlm.nih.gov/34707709/. DOI: 10.3892/etm.2021.10863.
[6] Damato B. Ocular treatment of choroidal melanoma in relation to the prevention of metastatic death – a personal view[J]. Prog Retin Eye Res, 2018, 66:187-199. DOI: 10.1016/j.preteyeres.2018.03.004.
[7] Calin GA, Croce CM. MicroRNA signatures in human cancers[J]. Nat Rev Cancer, 2006, 6(11):857-866. DOI: 10.1038/nrc1997.
[8] Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection[J/OL]. Proc Natl Acad Sci U S A, 2008, 105(30):10513-10518[2022-03-02]. https://pubmed.ncbi.nlm.nih.gov/18663219/. DOI: 10.1073/pnas.0804549105.
[9] Zhou WD, Shao L, Dong L, et al. Circulating microRNAs as quantitative biomarkers for diagnosis and prognosis of uveal melanoma[J/OL]. Front Oncol, 2022, 12:854253[2022-03-02]. https://pubmed.ncbi.nlm.nih.gov/35433428/. DOI: 10.3389/fonc.2022.854253.
[10] Li Q, Li B, Li Q, et al. Exosomal miR-21-5p derived from gastric cancer promotes peritoneal metastasis via mesothelial-to-mesenchymal transition[J/OL]. Cell Death Dis, 2018, 9(9):854[2022-03-03]. https://pubmed.ncbi.nlm.nih.gov/30154401/. DOI: 10.1038/s41419-018-0928-8.
[11] Ren W, Hou J, Yang C, et al. Extracellular vesicles secreted by hypoxia pre-challenged mesenchymal stem cells promote non-small cell lung cancer cell growth and mobility as well as macrophage M2 polarization via miR-21-5p delivery[J/OL]. J Exp Clin Cancer Res, 2019, 38(1):62[2022-03-03]. https://pubmed.ncbi.nlm.nih.gov/30736829/. DOI: 10.1186/s13046-019-1027-0.
[12] Jin XH, Lu S, Wang AF. Expression and clinical significance of miR-4516 and miR-21-5p in serum of patients with colorectal cancer[J/OL]. BMC Cancer, 2020, 20(1):241[2022-03-04]. https://pubmed.ncbi.nlm.nih.gov/32293319/. DOI: 10.1186/s12885-020-06715-6.
[13] Smit KN, Chang J, Derks K, et al. Aberrant microrna expression and its implications for uveal melanoma metastasis[J/OL]. Cancers (Basel), 2019, 11(6):815[2022-03-04]. https://pubmed.ncbi.nlm.nih.gov/31212861/. DOI: 10.3390/cancers11060815.
[14] Wang YC, Yang X, Wei WB, et al. Role of microRNA-21 in uveal melanoma cell invasion and metastasis by regulating p53 and its downstream protein[J]. Int J Ophthalmol, 2018, 11(8):1258-1268. DOI: 10.18240/ijo.2018.08.03.
[15] Liu YN, Shao L. Differential expression profile of microRNAs in different types of uveal melanoma[J]. Chin J Exp Ophthalmol, 2017, 35(9):778-785. DOI: 10.3760/cma.j.issn.2095-0160.2017.09.003.
[16] Simons M, Raposo G. Exosomes–vesicular carriers for intercellular communication[J]. Curr Opin Cell Biol, 2009, 21(4):575-581. DOI: 10.1016/j.ceb.2009.03.007.
[17] Zhang J, Li S, Li L, et al. Exosome and exosomal microRNA: trafficking, sorting, and function[J]. Genomics Proteomics Bioinformatics, 2015, 13(1):17-24. DOI: 10.1016/j.gpb.2015.02.001.
[18] Bhagirath D, Yang TL, Bucay N, et al. microRNA-1246 is an exosomal biomarker for aggressive prostate cancer[J]. Cancer Res, 2018, 78(7):1833-1844. DOI: 10.1158/0008-5472.CAN-17-2069.
[19] Xi Y, Nakajima G, Gavin E, et al. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples[J]. RNA, 2007, 13(10):1668-1674. DOI: 10.1261/rna.642907.
[20] Cortez MA, Bueso-Ramos C, Ferdin J, et al. MicroRNAs in body fluids–the mix of hormones and biomarkers[J]. Nat Rev Clin Oncol, 2011, 8(8):467-477. DOI: 10.1038/nrclinonc.2011.76.
[21] Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma[J]. Proc Natl Acad Sci U S A, 2011, 108(12):5003-5008. DOI: 10.1073/pnas.1019055108.
[22] Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins[J]. Nat Cell Biol, 2011, 13(4):423-433. DOI: 10.1038/ncb2210.
[23] Valihrach L, Androvic P, Kubista M. Circulating miRNA analysis for cancer diagnostics and therapy[J/OL]. Mol Aspects Med, 2020, 72:100825[2022-03-05]. https://pubmed.ncbi.nlm.nih.gov/31635843/. DOI: 10.1016/j.mam.2019.10.002.
[24] Ragusa M, Barbagallo C, Cirnigliaro M, et al. Asymmetric RNA distribution among cells and their secreted exosomes: biomedical meaning and considerations on diagnostic applications[J/OL]. Front Mol Biosci, 2017, 4:66[2022-03-05]. https://pubmed.ncbi.nlm.nih.gov/29046875/. DOI: 10.3389/fmolb.2017.00066.
[25] Fang YY, Tan MR, Zhou J, et al. miR-214-3p inhibits epithelial-to-mesenchymal transition and metastasis of endometrial cancer cells by targeting TWIST1[J]. Onco Targets Ther, 2019, 12:9449-9458. DOI: 10.2147/OTT.S181037.
[26] Zhou Z, Wu L, Liu Z, et al. MicroRNA-214-3p targets the PLAGL2-MYH9 axis to suppress tumor proliferation and metastasis in human colorectal cancer[J]. Aging (Albany NY), 2020, 12(10):9633-9657. DOI: 10.18632/aging.103233.
[27] Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function[J]. Nat Rev Immunol, 2002, 2(8):569-579. DOI: 10.1038/nri855.
[28] Zhao J, Wang F, Tian Q, et al. Involvement of miR-214-3p/FOXM1 axis during the progression of psoriasis[J]. Inflammation, 2022, 45(1):267-278. DOI: 10.1007/s10753-021-01544-6.
[29] Li D, Liu J, Guo B, et al. Osteoclast-derived exosomal miR-214-3p inhibits osteoblastic bone formation[J/OL]. Nat Commun, 2016, 7:10872[2022-03-05]. https://pubmed.ncbi.nlm.nih.gov/26947250/. DOI: 10.1038/ncomms10872.
[30] Tao W, Cao C, Ren G, et al. Circular RNA circCPA4 promotes tumorigenesis by regulating miR-214-3p/TGIF2 in lung cancer[J]. Thorac Cancer, 2021, 12(24):3356-3369. DOI: 10.1111/1759-7714.14210.
[31] Cagle P, Smith N, Adekoya TO, et al. Knockdown of microRNA-214-3p promotes tumor growth and epithelial-mesenchymal transition in prostate cancer[J/OL]. Cancers (Basel), 2021, 13(23):5875[2022-03-06]. https://pubmed.ncbi.nlm.nih.gov/34884984/. DOI: 10.3390/cancers13235875.
[32] Hu S, Chang J, Ruan H, et al. Cantharidin inhibits osteosarcoma proliferation and metastasis by directly targeting miR-214-3p/DKK3 axis to inactivate β-catenin nuclear translocation and LEF1 translation[J]. Int J Biol Sci, 2021, 17(10): 2504-2522. DOI: 10.7150/ijbs.51638