·Experimental Research·
Proteomic analysis of extraocular muscles from patients with thyroid-associated ophthalmopathy using a tandem mass tag
Meng Qingyu, Liang Shuting, Wang Yi, Wang Lejin, Zhao Mingwei, Wu Xi
Department of Ophthalmology, Peking University People’s Hospital, Eye Diseases and Optometry Institute, Beijing Key Laboratory of Diagnnosis and Therapy of Retinal and Choroid Diseases, College of Optometry, Peking University Health Science Center, Beijing 100044, China
Corresponding author: Wu Xi, Email: doctorwuxi@126.com
[Abstract] [View PDF in English] [View PDF in Chinese] [Read Full Text]
Objective This study aimed to identify the differentially expressed proteins (DEPs) in extraocular muscles from patients with restrictive strabismus having thyroid-associated ophthalmopathy (TAO) and those with concomitant esotropia by proteomic analysis using a tandem mass tag (TMT).
Methods Extraocular muscle samples from five patients with restrictive strabismus having TAO and five patients with concomitant esotropia were collected from August 2019 to December 2020. The patients received strabismus surgery. DEPs in the extraocular muscle samples were identified by quantitative proteomic analysis, and bioinformatic analysis was carried out using TMT. Fold change ≥1.2 or ≤0.83 and P value <0.05 were regarded as the thresholds to screen DEPs. Gene Ontology (GO) annotation, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, and protein–protein interaction (PPI) network analysis of DEPs were conducted through UniProt-Gene Ontology Annotation (GOA)and STRING database. This study protocol was approved by the ethics committee of Peking University People’s Hospital (No. 2021PHB058-001).
Results A total of 53 DEPs were identified, 34 of which were upregulated and 19 were downregulated. The biological processes in which DEPs mainly participated included response to stimulation, multicellular organismal process, metabolism, developmental process, intracellular signal transduction, and positive regulation of the biological process. DEPs were involved in pathways including focal adhesion, tight junction, regulation of action cytoskeleton, and apoptosis. Six key proteins identified using the PPI network included myosin heavy chain 2, myosin heavy chain 7, myosin regulatory light chain, α-actinin-2, fibrinogen alpha chain, and fibrinogen beta chain.
Conclusions DEPs were found in extraocular muscles from patients with restrictive strabismus having TAO and those with concomitant esotropia. Myosin, actinin, and filamin may be involved in the pathogenesis of TAO through the regulation of action cytoskeleton and focal adhesion.
[Key words] Proteomics; restrictive; strabismus; tandem mass tag; thyroid-associated ophthalmopathy
Fund program: National Key R&D Program of China (2020YFC2008200)
DOI: 10.3760/cma.j.cn115989-20210802-00442
Thyroid-associated ophthalmopathy (TAO) is an autoimmune disease 1,2. The tissues within the orbit, such as the extraocular muscles and orbital fat, as well as the thyroid tissue, are targeted by the immune response, leading to fibrosis of the extraocular muscles, loss of elasticity, and an increase in orbital soft tissue volume. This results in various eye features, such as different degrees of proptosis, incomplete eyelid closure, eyelid retraction, and compressive optic neuropathy3. TAO often involves the inferior and medial rectus muscles, resulting in restrictive hypotropia and esotropia 4. Patients with TAO often experience significant diplopia, which seriously affects their quality of life. The specific pathogenic mechanism of TAO is not yet clear 5,6. Tandem mass tag (TMT) is a peptide-labeling technology that uses multiple isotope labels to covalently bind with the amino groups of peptides in vitro. It can be used for the qualitative and quantitative analyses of proteins in 10 different samples at the same time. TMT has the advantages of accurate quantification, good reproducibility, and high sensitivity, and is widely used in differential protein expression analysis7. This study used protein isotope–labeling quantitative analysis combined with liquid chromatography–tandem mass spectrometry to determine the secondary restrictive strabismus extraocular muscles and common strabismus extraocular muscles in TAO, screen for differentially expressed proteins (DEPs), and perform bioinformatics analysis of the functions of these proteins to identify the potential biomarkers involved in the occurrence and development of TAO. This study provided a research foundation for further investigating the pathogenesis of TAO.
1 Materials and Methods
1.1 Sample collection
Five patients diagnosed with TAO and concurrent restrictive strabismus who underwent strabismus correction surgery were recruited from the Department of Ophthalmology, Peking University People’s Hospital, between August 2019 and December 2020. During surgery, samples of the extraocular muscles were collected. The control group consisted of five patients diagnosed with concomitant esotropia who underwent strabismus correction surgery during the same period, and extraocular muscle samples were collected from them. This study protocol was approved by the ethics committee of Peking University People’s Hospital (Approval No. 2021PHB058-001).
1.2 TMT labeling and mass spectrometry detection
1.2.1 Protein digestion All muscle samples were added to the lysis buffer, and two steel beads were placed in a grinder for tissue lysis. The mixture was centrifuged at 20,000g for 30 min at 4°C, and the supernatant was collected. Trichloroacetic acid (TCA)-acetone (four times the volume of the supernatant) was added, and the mixture was precipitated at -20°C for at least 2 h. The mixture was centrifuged at 20,000g for 30 min at 4°C, and then three volumes of pure acetone at -20°C were added. This step was repeated three times, and the precipitate was washed. The lysate was added to the precipitate and sonicated for 5 min to help in dissolution. The sonication conditions were as follows: pulse on 2 s, pulse off 3 s, power 180 W, and temperature 4°C. The lysate was then centrifuged at 20,000g for 30 min to collect the supernatant. The protein concentration of the sample was determined using the Bradford method. First, 30 μg of the lysate was taken from each sample, added to a 10K ultrafiltration tube, and centrifuged at 14,000g for 40 min at 4°C. The filtrate was discarded, and 200 μL of 50% (vol/vol) tetraethyl ammonium bromide (TEAB) was added, followed by centrifugation at 14,000g for 40 min at 4°C. The process was repeated twice, and 1 μg/μL trypsin was added to the samples, with 3.3 μg of the enzyme added per 100 μg of the protein substrate. The samples were incubated in a 37°C water bath for 24 h. The digestion solution was freeze-dried, and then 25 μL of the 100 mmol/L TEAB was added to each tube to resuspend the peptide segments.
1.2.2 TMT labeling of peptide segments The labeling reagent was taken out at 4°C and equilibrated to room temperature. Then, 41 μL of acetonitrile was added to each tube containing the labeling reagent and vortexed for 1 min. The mixed labeling reagent was added to the peptide segments, with different samples labeled with different sizes of isotopes. The mixture was vortexed and left at room temperature for 1 h. Then, 8 μL of 5% hydroxylamine was added and left at room temperature for 15 min. The sample was mixed and dried under a vacuum.
1.2.3 Reversed-phase high-performance liquid chromatography Reversed-phase high-performance liquid chromatography (RP-HPLC) separation was performed using a Phenomenex Luna SCX 250 × 4.60 mm 100 Ǻ column. The mobile phase consisted of 95% H2O + 5% acetonitrile (water phase) and 95% acetonitrile + 5% H2O (organic phase), both adjusted to pH 9.8 with ammonium hydroxide. The sample, which was dried as mentioned in Section 1.2.2, was dissolved in 1 mL of water and centrifuged at 15,000g for 10 min at 4℃. The supernatant was subjected to a preseparated chromatogram using a set gradient elution. Elution was initiated at 1 min after peak detection, and the fractions were collected every 30 s until 130 min, which were combined into 10 fractions and vacuum dried.
1.2.4 Mass spectrometry detection and differential protein screening Mass spectrometry detection and differential protein screening were performed using a high-precision liquid chromatography–mass spectrometry system (Q-Exactive-Orbitrap; Thermo Fisher Scientific, Waltham, MA, 02454, USA) to detect peptide signals. The mass spectrum signals were acquired and converted into an mgf file. The Mascot software (MASCOT 2.3.01) was used for qualitative retrieval and quantitative analysis. The Swiss Human species database was used for protein identification with a false discovery rate of <1% as the filtering criterion. The relative quantification of peptides was based on ion abundance, and that of proteins on the relative quantification of peptides. Differential protein screening was performed based on a fold change of greater than 1.2 or less than 0.83 and P <0.05 between the two groups for the same protein.
1.3 Bioinformatics analysis
1.3.1 Protein annotation method DEPs were annotated using the UniProt-GOA database for Gene Ontology (GO) analysis. The software Proteome Discoverer 1.4 and the corresponding InterPro domain database were used for protein domain annotation based on protein sequence algorithms.
1.3.2 Protein functional enrichment Protein functional enrichment was divided into three parts: GO enrichment analysis, pathway enrichment analysis, and protein domain enrichment analysis. The protein GO annotations were divided into three main categories: biological processes, cellular components, and molecular functions. Fisher’s exact test was used to evaluate DEPs against the background of identified proteins. The GO enrichment tests with P < 0.05 were considered to have statistical significance. The InterPro database was used to analyze the enrichment of functional domains of DEPs. Enrichment tests for domain units with P < 0.05 were considered to have statistical significance.
1.3.3 Protein clustering analysis First, the quantitative information of the target protein set was normalized to the range (-1, 1). Then, the ComplexHeatmap package in the R software (version 3.4) was used to perform two-dimensional clustering on the protein expression levels. Finally, a hierarchical clustering heat map was generated.
1.3.4 Protein–protein interaction network analysis Using the STRING (V.10.5) protein network interaction database as a reference, the differential protein database IDs or protein sequences from different comparison groups were compared to extract differential protein interaction relationships with a confidence score >0.7 (high confidence). The software CytoScape (version 3.2.1) was used to generate the protein–protein interaction (PPI) network connections.
2 Results
2.1 Differential protein screening results
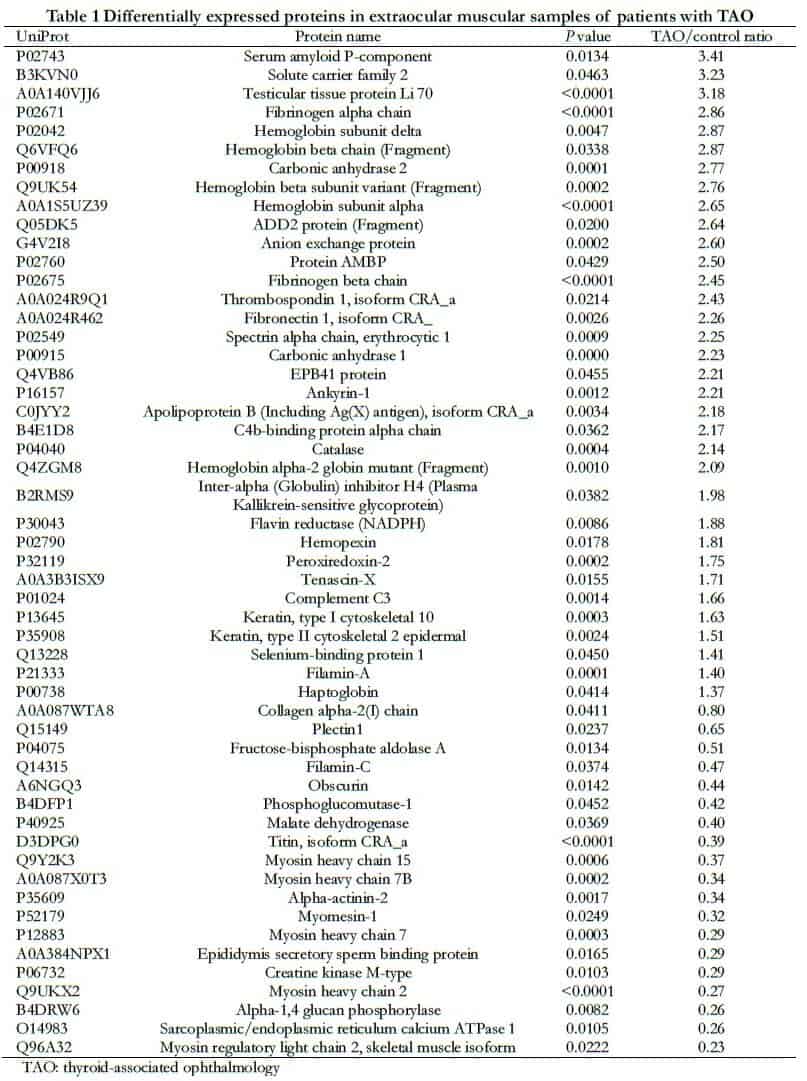
A total of 24,265 spectrum segments were identified, with 9140 peptide segments, 8011 unique peptide segments, and 1697 proteins. A total of 53 DEPs were identified in the TAO and control groups, including 34 upregulated and 19 downregulated proteins (Table 1). The volcano plot of protein quantification is shown in Figure 1.
2.2 Functional informatics analysis of DEPs
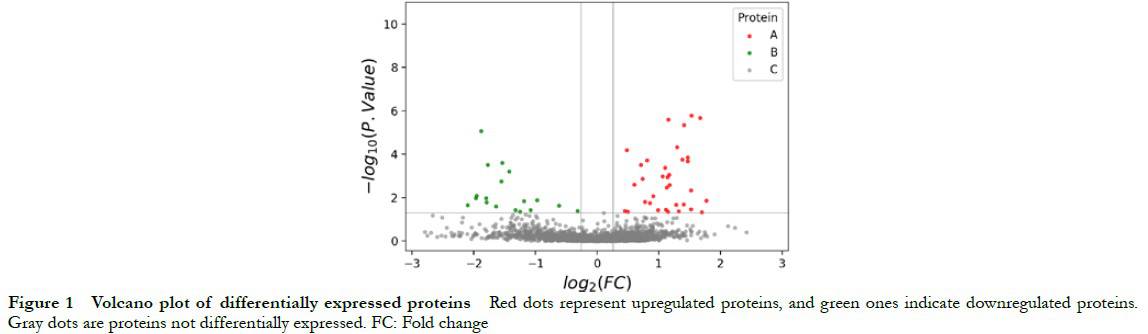
2.2.1 GO analysis of DEPs From the perspective of molecular function, DEPs had binding and transportation functions. In terms of biological processes, these proteins mainly participated in processes such as response to stimuli, multicellular organism processes, metabolic processes, developmental processes, intracellular signal transduction, and positive biological regulation processes. In terms of cellular composition, these proteins were mainly located in the intracellular region, cytoplasm, extracellular region, and cytosol (Fig. 2).
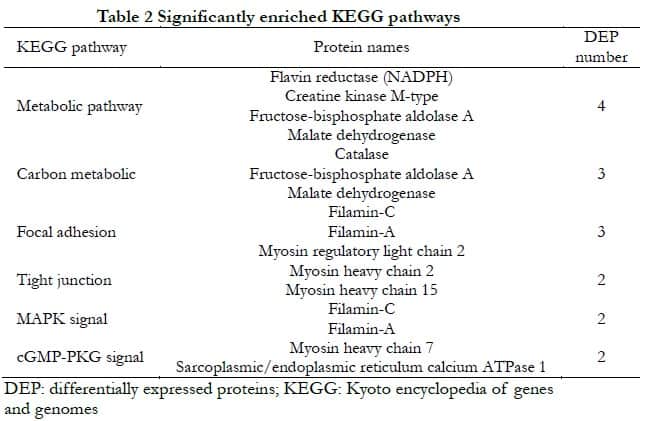
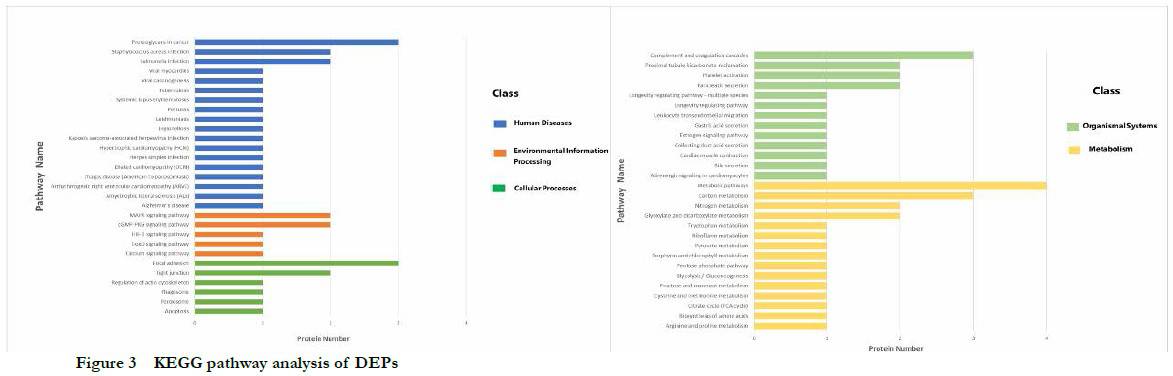
2.2.2 KEGG pathway enrichment analysis of DEPs The cell process classification showed that DEPs mainly participated in estrogen signaling pathway, myocardial cell adrenal signaling, myocardial contraction, leukocyte chemotaxis, gastric acid secretion, lifespan regulation pathway, platelet, and pancreatic secretion. Environmental information processing classification showed that DEPs mainly participated in glycation, amino acid nuclear magnetic resonance, acetic acid metabolism, TCA metabolic cycle, vitamin B12 metabolism, nitrogen metabolism, and carbon metabolism. Human disease classification showed that DEPs mainly participated in Alzheimer’s disease, hypertrophic cardiomyopathy, dilated cardiomyopathy, viral myocarditis, and so forth. Metabolic classification showed that DEPs mainly participated in the calcium pathway, hypoxia-inducible factor-1 (HIF-1) signaling pathway, Forkhead box O (FoxO) signaling pathway, cyclic guanosinc monophosphate-protein kinase G (cGMP-PKG) signaling pathway, and mitogen-activated protein kinase (MAPK) signaling pathway. Biological system classification showed that DEPs mainly participated in pathways such as focal adhesion, tight junction, cytoskeleton regulation, phagosome, and apoptosis (Fig. 3). The significantly enriched pathways and their DEPs are shown in Table 2.
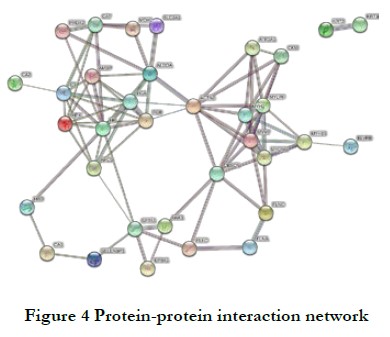
2.2.3 Regulation network of PPIs A complex regulation network consisting of 34 nodes and 85 edges was formed, with an average node degree of 5 and a clustering coefficient of 0.524 (Fig. 4). Six key proteins were identified: myosin heavy chain (MYH2 and MYH7), myosin light chain (MYLPF), alpha-actinin-2 (ACTN2), and fibrinogen alpha and beta chains (FGA and FGB).
3 Discussion
This study proposed that the target of TAO was in the orbital fibroblasts. The thyroid-stimulating hormone receptor (TSHR) was considered to be the main autoantigen in Graves’ disease, and the expression of TSHR mRNA in orbital fibroblasts was elevated in patients with TAO compared with normal controls 8,9. The insulin-like growth factor 1 receptor (IGF-1R) is a transmembrane tyrosine kinase receptor that can form physical and functional complexes with TSHR, activate downstream signaling pathways including the cyclic adenosine monophosphate/protein kinase A (cAMP/PKA) and phosphatidylionsitol 3-kinase (PI3K) pathways, and promote the activation of orbital fibroblasts10,11. Activated orbital fibroblasts show increased proliferation and activity, can produce inflammatory mediators, differentiate into adipocytes and myofibroblasts, and produce excessive extracellular matrix components1,6. However, the pathogenesis of TAO is still unclear. In this study, TMT proteomic analysis was used to determine and analyze DEPs in the restricted lateral rectus muscle of patients with TAO having esotropia and the coexisting medial rectus muscle of patients with TAO having exotropia to lay the experimental foundation for further exploring the pathogenesis of TAO.
The functional annotation analysis revealed that DEPs were mainly involved in stress response, metabolic processes, and cell signal transduction. The KEGG pathway analysis revealed that DEPs were mainly involved in the MAPK signaling pathway, FoxO signaling pathway, cGMP-PKG signaling pathway, focal adhesion pathway, cell cytoskeleton regulation, and cell apoptosis. Combined with PPI analysis, potential key proteins in TAO were identified, including MYH2 and MYH7, MYLPF, ACTN2, and fibrinogen.
Myosin is the most abundant protein in skeletal muscle and a major component of thick filaments. It plays an important role in muscle contraction, metabolism, and cell structure. Myosin comprises one pair of heavy chains and two pairs of light chains, including one pair of myosin essential light chains and one pair of MYLPF12. The essential light chain stabilizes the structure of the heavy chain, while the regulatory light chain MYLPF regulates the activity of myosin. Previous studies found that the function of MYH subtypes and regulatory light chains was related to striated muscle contraction13,14. An abnormal expression of MYH2, MYH7, and MYLPF in patients with TAO may affect the contraction strength and speed of extraocular muscle fibers, leading to eye muscle dysfunction.
Actin is another important component of the cell skeleton and forms contractile protein in muscle fibers by combining with myosin15. ACTN2 is a muscle actin cross-linking protein located in the Z-disk of the sarcomere. It acts as a link between antiparallel actin filaments and enhances the stability of the Z-disk by binding to the N-terminus of actin. ACTN2 plays an important role in stabilizing cell adhesion and regulating cell shape and cell movement. Mutations in the ACTN2 gene can lead to different types of congenital myopathy16. This study found that the level of ACTN2 was downregulated in the extraocular muscles of patients with TAO, indicating a decrease in ACTN2 function, which may disrupt the stability of the Z-disk and lead to the occurrence of strabismus.
Fibrinogen is a plasma protein coagulation factor synthesized and secreted by liver cells, which participates in the body’s coagulation system and is involved in infection and inflammation processes17. Thyroid disease is closely related to the coagulation system18. Previous studies showed that the fibrinogen level was negatively correlated with thyroid-stimulating hormone levels and positively correlated with the levels of free thyroxine T3 and T419. This study found that the alpha and beta chains of fibrinogen were upregulated in the extraocular muscle tissue of patients with TAO, which might be related to the patient’s thyroid hormone levels.
The actin-binding protein filamin belongs to the family of actin-binding proteins and is an important cross-linker of actin filaments 20. Filamin plays a crucial role in regulating the mechanical strength and plasticity of the cellular skeleton by binding to actin. Mutations in the filamin (FLN) gene that encodes for filamin can cause developmental abnormalities in multiple organs 21,22. Recent studies showed that filamin played a key role in cell adhesion and migration to the extracellular matrix 21,23. In combination with the KEGG pathway enrichment analysis in this study, both Filamin-A and Filamin-C were found to be involved in the local adhesion pathway and the MAPK pathway (Table 2). Previous studies by domestic scholars showed an upregulation of adhesion molecules in the TAO extraocular muscles, indicating a role of cell adhesion pathways in TAO pathogenesis24. These findings combined with the results of this study suggested that filamin might be involved in the occurrence and development of TAO by affecting cell adhesion processes.
Previous studies confirmed the involvement of TSHR, IGF-1R, interleukin (IL)-6, and IL-10 in the pathogenesis of TAO 25,26. However, in this study, no significant difference was observed in the expression of these molecules between the TAO and control groups in extraocular muscle specimens. This might be because the number of TSHR immunostaining–positive cells and TSHR mRNA expression were higher in the early stage of TAO, but decreased with prolonged disease duration in extraocular muscle samples. In addition, compared with patients with active Graves’ disease, those with nonactive Graves’ disease had lower mRNA expression levels of pro-inflammatory cytokines, such as IL-1β, IL-6, IL-8, and IL-10, in their orbital fat/connective tissue 27,28. All the patients included in this study were in the nonactive phase of TAO; therefore, no significant differences were observed in the protein expression levels of TSHR, IGF-1R, IL-6, and IL-10.
This study used TMT-based liquid chromatography–tandem mass spectrometry to quantify DEPs in the secondary restrictive and concomitant strabismus medial rectus muscles of patients with TAO. A total of 53 DEPs were identified, including 34 upregulated and 19 downregulated proteins. The KEGG pathway enrichment analysis revealed that the DEPs were mainly involved in pathways related to local adhesion, tight junctions, cytoskeleton regulation, phagosomes, and apoptosis. Based on these results, further cellular and molecular studies on protein function and signal transduction pathways are needed to elucidate the precise pathogenesis of TAO and identify potential therapeutic targets.
Conflict of interests The authors declare no conflicts of interest.
Authors’ contribution Qingyu Meng participated in the experimental design, implementation, data collection, data interpretation, and drafting of the manuscript. Shutin Liang participated in the implementation of the research, data collection, manuscript revision, and statistical analysis. Yi Wang participated in data collection, manuscript review, and research guidance. Lejin Wang participated in data collection, manuscript revision, and research guidance. Mingwei Zhao participated in the experimental design, manuscript review, and research guidance. Xi Wu participated in the experimental design, implementation, supervision of the research progress, and manuscript review and revision.
References
- Bahn RS. Graves’ ophthalmopathy. N Engl J Med. 2010;362(8):726-738.
- Ginsberg J. Diagnosis and management of Graves’ disease. CMAJ. 2003;168(5):575-585.
- Subekti I, Pramono LA. Current Diagnosis and Management of Graves’ Disease. Acta Med Indones. 2018;50(2):177-182.
- Sahli E, Gunduz K. Thyroid-associated Ophthalmopathy. Turk J Ophthalmol. 2017;47(2):94-105.
- Taylor PN, Zhang L, Lee RWJ, et al. New insights into the pathogenesis and nonsurgical management of Graves orbitopathy. Nat Rev Endocrinol. 2020;16(2):104-116.
- Wang ZM, Wang ZY, Lu Y. The role of cell mediated immunopathogenesis in thyroid-associated ophthalmopathy. Int J Ophthalmol. 2019;12(7):1209-1214.
- Moulder R, Bhosale SD, Goodlett DR, Lahesmaa R. Analysis of the plasma proteome using iTRAQ and TMT-based Isobaric labeling. Mass Spectrom Rev. 2018;37(5):583-606.
- Menconi F, Marcocci C, Marino M. Diagnosis and classification of Graves’ disease. Autoimmun Rev. 2014;13(4-5):398-402.
- Douglas RS, Afifiyan NF, Hwang CJ, et al. Increased generation of fibrocytes in thyroid-associated ophthalmopathy. J Clin Endocrinol Metab. 2010;95(1):430-438.
- Smith TJ, Janssen J. Insulin-like Growth Factor-I Receptor and Thyroid-Associated Ophthalmopathy. Endocr Rev. 2019;40(1):236-267.
- Dai JL, He WM, Luo MQ. Promoting effects of insulin-like growth factor-1 on proliferation of orbital fibroblasts derived from thyroid associated ophthalmopathy[J]. Chin J Exp Ophthalmol, 2017, 35(9):805-810. DOI: 10.3760/cma.j.issn.2095-0160.2017.09.008.
- Pette D, Staron RS. Myosin isoforms, muscle fiber types, and transitions. Microsc Res Tech. 2000;50(6):500-509.
- Wu L, Zhang S, Li X, et al. Integrative transcriptomics and proteomic analysis of extraocular muscles from patients with thyroid-associated ophthalmopathy. Exp Eye Res. 2020;193:107962.
- Hoh JFY. Myosin heavy chains in extraocular muscle fibres: Distribution, regulation and function. Acta Physiol (Oxf). 2021;231(2):e13535.
- Sweeney HL, Houdusse A, Robert-Paganin J. Myosin Structures. Adv Exp Med Biol. 2020;1239:7-19.
- Ranta-Aho J, Olive M, Vandroux M, et al. Mutation update for the ACTN2 gene. Hum Mutat. 2022;43(12):1745-1756.
- Litvinov RI, Pieters M, de Lange-Loots Z, Weisel JW. Fibrinogen and Fibrin. Subcell Biochem. 2021;96:471-501.
- Elbers LPB, Fliers E, Cannegieter SC. The influence of thyroid function on the coagulation system and its clinical consequences. J Thromb Haemost. 2018;16(4):634-645.
- Ellervik C, Mora S, Kus A, et al. Effects of Thyroid Function on Hemostasis, Coagulation, and Fibrinolysis: A Mendelian Randomization Study. Thyroid. 2021;31(9):1305-1315.
- Stossel TP, Condeelis J, Cooley L, et al. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2(2):138-145.
- Zhou J, Kang X, An H, Lv Y, Liu X. The function and pathogenic mechanism of filamin A. Gene. 2021;784:145575.
- Nakamura F, Stossel TP, Hartwig JH. The filamins: organizers of cell structure and function. Cell Adh Migr. 2011;5(2):160-169.
- Lamsoul I, Dupre L, Lutz PG. Molecular Tuning of Filamin A Activities in the Context of Adhesion and Migration. Front Cell Dev Biol. 2020;8:591323.
- Peng J, Xu XL. Expression of intercellular adhesion molecule in extraocular muscles with thyriod associated ophthalmopathy[J]. Chin J Exp Ophthalmol, 2007, 25(7):522-525. DOI: 10.3760/cma.j.issn.2095-0160.2007.07.014..
- Huang D, Xu N, Song Y, Wang P, Yang H. Inflammatory cytokine profiles in the tears of thyroid-associated ophthalmopathy. Graefes Arch Clin Exp Ophthalmol. 2012;250(4):619-625.
- Shen J, Li Z, Li W, et al. Th1, Th2, and Th17 Cytokine Involvement in Thyroid Associated Ophthalmopathy. Dis Markers. 2015;2015:609593.
- Boschi A, Daumerie C, Spiritus M, et al. Quantification of cells expressing the thyrotropin receptor in extraocular muscles in thyroid associated orbitopathy. Br J Ophthalmol. 2005;89(6):724-729.
- Wakelkamp IM, Bakker O, Baldeschi L, Wiersinga WM, Prummel MF. TSH-R expression and cytokine profile in orbital tissue of active vs. inactive Graves’ ophthalmopathy patients. Clin Endocrinol (Oxf). 2003;58(3):280-287.