Specific expression of transmembrane protein, TMEM26, in retinas and its association with primary open-angle glaucoma
Yin Yi, Mao Yao, Yang Zhenglin, Huang Lulin
School of Medicine, University of Electronic Science and Technology of China, Department of Medical Genetics, Sichuan Academy of Medical Sciences & Sichuan Provincial People’s Hospital, Sichuan Provincial Key Laboratory for Human Disease Gene Study, Chengdu 610072, China
Corresponding author: Huang Lulin, Email: huangluling@yeah.net
Abstract [View PDF in English] [View PDF in Chinese] [Read Full Text]
Objective
To predict the transmembrane structure of transmembrane protein 26 (TMEM26), observe its expression in human retina and mouse retina, and investigate the relationship between it and primary open-angle glaucoma (POAG).
Methods
The transmembrane structure of TMEM26 in human and mouse was obtained by inputting its amino acid sequences into the transmembrane protein structure prediction software, MemBrain.The expression and location of TMEM26 in human and mouse retinas were observed through frozen retinal sections stained with anti-TMEM26 antibody, which came from a human donor and five SPF-grade C57BL/6 mice.The possible function of TMEM26 gene and its influence on eyes were inferred on the basis of the specific expression of TMEM26 in retina.The single nucleotide polymorphism mutation of TMEM26 gene was searched in literature related to ocular diseases.The use and care of animals complied with the Regulations on the Management of Experimental Animals.This research protocol was approved by an Ethics Committee of Sichuan Provincial People’s Hospital (No.2019-36).
Results
Both human and mouse TMEM26 were eight transmembrane proteins with similar eight hydrophobic transmembrane domains, four hydrophilic cytoplasmic domains and five hydrophilic extracellular membrane domains.Small differences in the number of amino acid residues in the domains of TMEM26 were found.In both human and mouse retina, TMEM26 gene was only specifically expressed in the outer plexiform layer (OPL)and inner plexiform layer (IPL). TMEM26 was weakly associated with POAG in a published data.
Conclusions
TMEM26 is a multi-pass transmembrane protein, mainly expressed in IPL and OPL of the retina. TMEM26 gene is weakly related to POAG.
Key words:
As functional proteins, membrane proteins are important in living organisms, and are involved in cell proliferation and differentiation, growth and metabolism, material exchange, and energy transformation[1]. Membrane proteins can be classified into membrane-integrated, membrane-anchored, and extrinsic membrane proteins. Transmembrane proteins, the membrane-integrated proteins, are responsible for signal transduction and transmembrane transport of small molecule substances or ions[2]. Because there is at least one segment of transmembrane structure in transmembrane proteins, the transmembrane protein structure includes the transmembrane, extracellular, and cytoplasmic domains, which are important for the function of these proteins. Located on chromosome 10, transmembrane protein 26 (TMEM26) is a 6-exon gene expressed in both human and mouse, which encodes multi-pass transmembrane proteins with a relative molecular mass of about 41,600[3]. Currently, it has been verified that TMEM26 is expressed in mouse embryos and is regulated by time of development and space[3]; it is expressed in breast cancer cells, and its protein structure affects the response to drug therapy[4]. It is also expressed in airway epithelium and lung parenchymal cells, and is a candidate gene related to decreased lung function and the pathogenesis of chronic obstructive pulmonary disease[5]. Moreover, TMEM26 has been identified as a surface marker for beige adipocytes[6-7]. Although TMEM26 is expressed in various tissues and cells, its specific functions, especially in the eye, remain unclear. Glaucoma is a major cause of blindness, and commonly includes chronic angle-closure glaucoma, primary open-angle glaucoma (POAG), and exfoliation glaucoma. The loss of retinal ganglion cells (RGCs) is a key pathogenesis of glaucoma[8-9]. The inner plexiform layer (IPL) is composed of a fibrous reticular layer formed by RGC dendrites, bipolar cell axons, and amacrine cells. Its thickness is a potential biomarker for POAG[10]. This study therefore aimed to characterize the expression of TMEM26 in the retina and its association between variations of TMEM26 gene POAG, based on the reported data of this study.
1 Materials and Methods
1.1 Materials
1.1.1 Material sources. Five healthy SPF C57BL/6 mice (21 weeks old, 25−31 g) were provided by Jiangsu GemPharmatech, Nanjing, China), and their feeding and use conformed to the Regulations on the Management of Experimental Animals. One human eyeball was from a cadaver donor. This study was approved by the Ethics Committee of Sichuan Provincial People’s Hospital [Approval No.: LS (Y) 2019 No. 36], and the families of the donors provided written consent for the use of human specimens.
1.1.2 Main reagents and instruments. The following reagents and instruments were used: 4% paraformaldehyde universal tissue fixative solution (Biosharp, Beijing, China); 30% sucrose solution (Beijing Solarbio Science & Technology, Beijing, China); optimal cutting temperature (OCT) cryo embedding medium (Sakura Finetek, Torrance, CA, USA); rabbit anti-TMEM26 polyclonal antibody (TA330777; Origene, San Diego, CA, USA); Isolectin B4 594 (MKbio, Yamaguchi, Japan); 0.25% Triton X-100 (Beijing Solarbio Science & Technology); 5% goat serum, Alexa Fluor 488-labeled goat anti-rabbit fluorescent secondary antibody, 4′,6-diamidino-2-phenylindole (DAPI) and phosphate-buffered saline (PBS) (Shanghai Beyotime Biotechnology, Shanghai, China); a dissecting microscope (SZX10; Olympus, Tokyo, Japan); microdissecting instruments (Shenzhen Biotechnology, Shenzhen, China); a freezing microtome (BK-2318; Biobase Biodustry (Shandong) Co., Ltd, Jinan, China); a constant temperature oven (DHP-9052; Shanghai Hecheng Instrument Manufacturing, Shanghai, China); and a laser scanning confocal microscope (LSM800; Carl Zeiss, Jena, Germany).
1.2 Methods
1.2.1 Prediction of transmembrane protein structures. The amino acid sequences of human and mouse TMEM26 were obtained from the UCSC website (http://genome.ucsc.edu/), and then inputted into MemBrain 3.0 (http://www.csbio.sjtu. edu.cn/bioinf/MemBrain/).
1.2.2 Observation of expression and localization of TMEM26 in human and mouse retinas by immunofluorescence staining. The cornea was removed using a dissecting microscope, and the eyeball was fully immersed in 4% paraformaldehyde. After 2 h of fixation, residual paraformaldehyde was removed using phosphate-buffered saline (PBS) solution. The lens was then gently removed using tweezers and a dissecting microscope. The eyeball without the lens was dehydrated with 30% sucrose for 2 h, embedded with OCT embedding medium, and stored in a refrigerator at -80°C, followed by frozen sectioning. The sections were then baked in an oven at 37°C for 1 h to completely fix them on the slides. The position of the sections was circled with an immunohistochemical pen, and the sections were blocked with PBS containing 5% goat serum and 0.25% Triton X-100 for 1 h, then incubated with rabbit anti-TMEM26 polyclonal antibody (1:200) in the dark at 4°C overnight with Alexa Fluor 488-labeled goat anti-rabbit secondary antibody (1:500), and then with DAPI in the dark for 2 h. After washing with PBS, the sections were observed and photographed using a laser scanning confocal microscope.
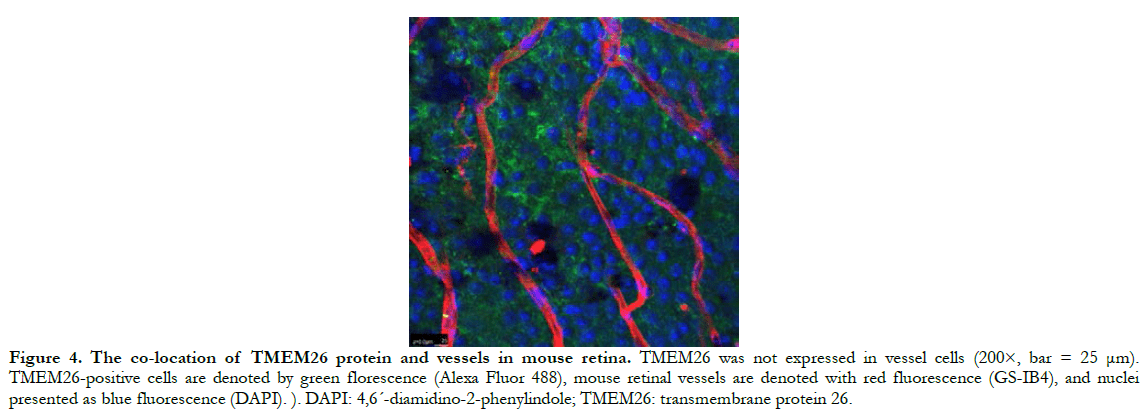
1.2.3 Co-localization of TMEM26 and vessels by double immunofluorescence staining. TMEM26 immunofluorescence staining was performed in accordance with Section 1.2.2, and retinal vascular endothelial cells were marked by Isolectin B4 staining. The cells were stained with Isolectin B4 (1:200) in the dark at 4°C overnight, washed with PBS for 10 min, mounted, and observed, and photographed using a laser scanning confocal microscope. DAPI, TMEM26, and retinal vessels were labeled blue, green, and red, respectively.
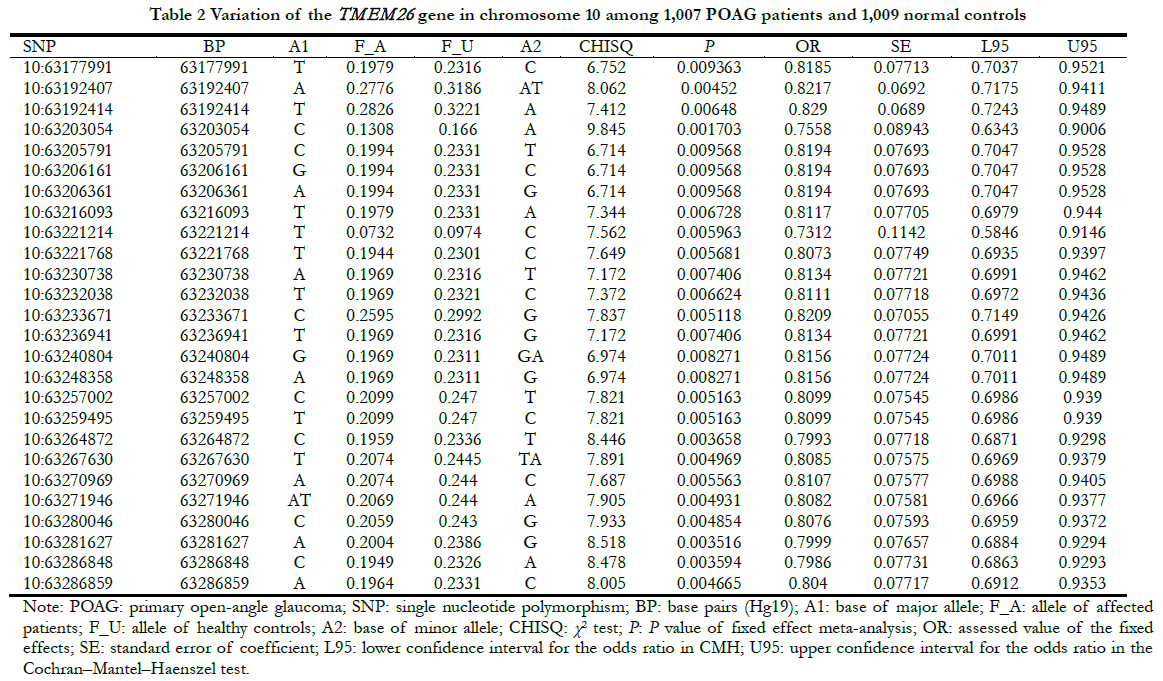
1.2.4 Single nucleotide polymorphism (SNP) analysis of TMEM26. Based on previous data of our group [11], the SNP variation of TMEM26 was analyzed in 1,007 POAG patients and 1,009 normal controls. The data were then summarized to identify significant differences (p < 0.01).
1.3 Data processing
TMEM26 gene and protein information was obtained from NCBI (https://www.ncbi.nlm.nih.gov/) and the UCSC website. The transmembrane protein structure was predicted using MemBrain 3.0. DNAMAN (https://www.lynnon.com/) was used for amino acid sequence alignment, and transmembrane protein images were edited by AI.
2 Results
2.1 The transmembrane structure of TMEM26
According to the UCSC website, there were 368 amino acid residues in human TMEM26 and 366 amino acid residues in mouse TMEM26. The amino acid sequences of TMEM26 from the two sources were compared, showing a homology of 69.46%. The predicted transmembrane structure of human and mouse TMEM26 is shown in Figure 1, and the amino acid sequences are shown in Table 1. Human TMEM26 contained eight hydrophobic transmembrane domains, four hydrophilic cytoplasmic domains, and five hydrophilic extracellular domains, and had a predicted hydrophilic cytoplasmic structure connected by two helical transmembrane structures, with only two amino acid residues, which was a small cytoplasmic domain. Similar to the predicted transmembrane structure of human TMEM26 (Figure 1 and Table 1), mouse TMEM26 also contained eight predicted transmembrane domains, five hydrophilic extracellular domains, and four hydrophilic cytoplasmic domains. Both human and mouse TMEM26 had polypeptide chain N-termini and C-termini outside the cell membrane, and their transmembrane structures were similar, with similar transmembrane, extracellular, and cytoplasmic domains. However, the numbers of amino acid residues of the domains were slightly different. The predicted two “back-to-back” helical transmembrane structures in human TMEM26 consisted of 29 amino acid residues of E2-V30 and 25 amino acid residues of E33-K57, while those of mouse TMEM26 consisted of 30 amino acid residues of E2-K31 and 24 amino acid residues of H34-K57. In addition, the number of amino acid residues showed the greatest difference in the extracellular domain at the terminal of the polypeptide, i.e., there were two more amino acid residues in human TMEM26 than mouse TMEM26.
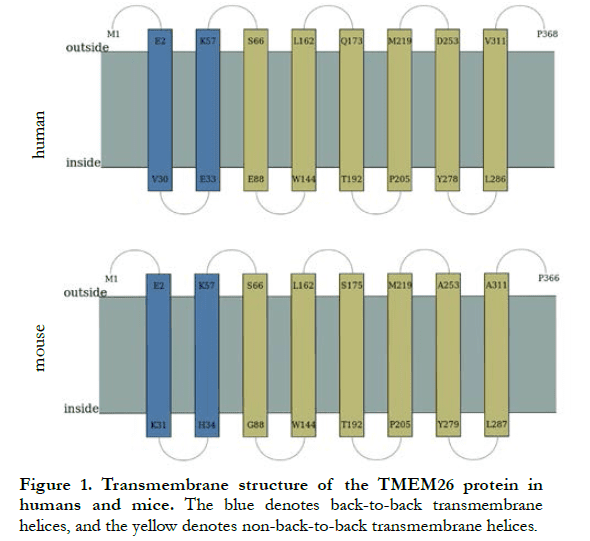
2.2 Expression of TMEM26 in retinas
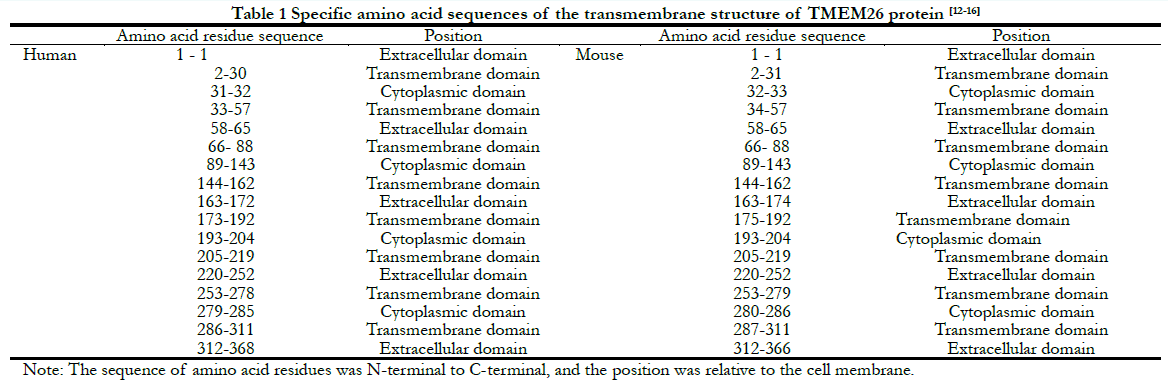
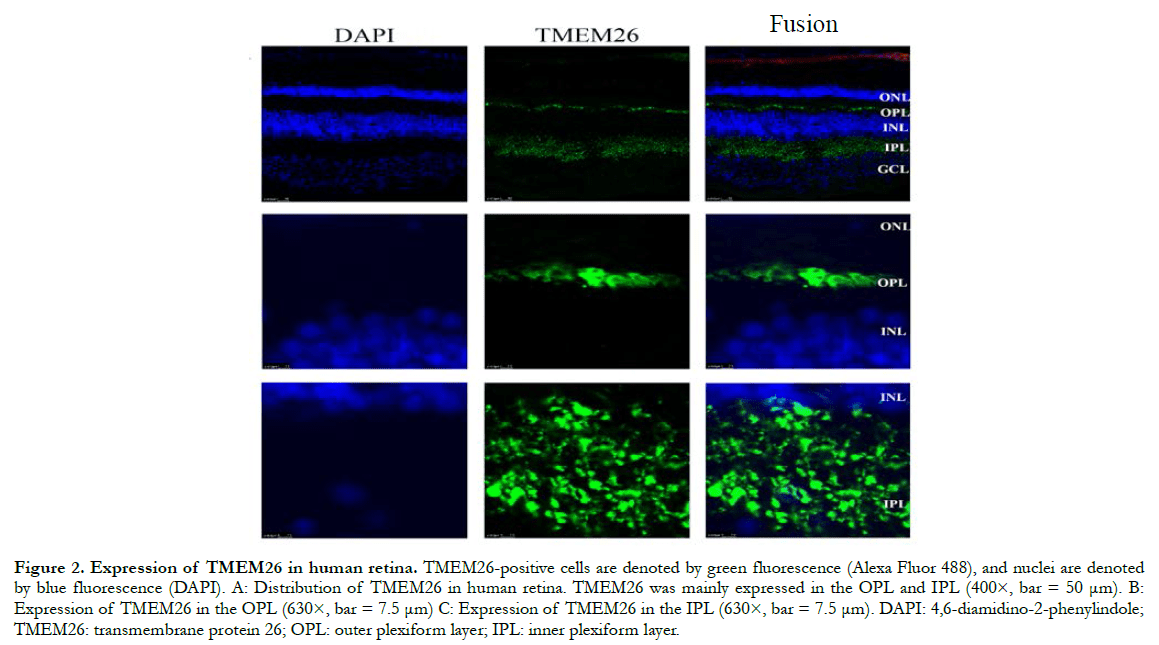
In human retinas, the TMEM26 protein was mainly specifically expressed in the outer plexiform layer (OPL) and inner plexiform layer (IPL) of the retina, rather than in the nucleus (Figure 2). In a similar manner, in the mouse retinas, TMEM26 was expressed primarily in the OPL and IPL of the retina, rather than in the nucleus (Figure 3). In addition, isolectin B4 staining showed that TMEM26 was not expressed in vascular cells (Figure 4).
2.3 Association between TMEM26 SNP and POAG
TMEM26 was mainly expressed in the IPL and OPL of the retina, showing a specific expression pattern. To determine whether the presence of TMEM26 was associated with eye disease, the association between TMEM26 and POAG was determined, based on the published data of the Genome-wide Association Study (GWAS) [11]. The results showed that among 1,007 POAG patients with high intraocular pressure and 1,009 controls, 26 TMEM26 SNP sites were weakly correlated with POAG (p = E-3), with the 10:63203054 site having the strongest signal (p = 0.0017, odds ratio = 0.76), suggesting that it may have a weak protective effect on POAG (Table 2).
3 Discussion
TMEM26, a member of the transmembrane protein family, is located on the cell membrane. Based on immunohistochemical staining, TMEM26 is expressed on the cytoplasmic side rather than the nucleus. Its expression is thought to occur in synapses, which secrete neurotransmitters. Membrane proteins, especially transmembrane proteins, are important for material transport and receptor recognition. The transmembrane structure of multi-pass transmembrane proteins can form material transport channels, while the hydrophilic domains of transmembrane proteins exert regulatory effects in channel switching and protein recruitment. Studying the transmembrane protein structure is therefore of great significance for understanding and identifying the function of transmembrane proteins. Comparisons of the transmembrane structures of human and mouse TMEM26 can provide the basis for animal experiments, and can provide possible etiologies of human eye diseases. Slight difference in the structures of human and mouse TMEM26 may suggest differences in the regulatory effects and material transport, but its specific significance remains to be determined. There have been few studies on the structure and function of TMEM26. As a protein-coding gene, the 6-exon mRNA of about 6 kb in TMEM26 is the most common transcript. In addition, there are other transcripts. For example, an 82 bp exon 2a is present between exon 2 and exon 3 of TMEM26 in mice, and the exon 2a insert leads to a frameshift mutation. Finally, early stop codons have been introduced into exon 3, resulting in early termination of translation and abnormal TMEM26 gene expression [3].
The presence or absence of N-terminal signal peptides and the number of C-terminal domains will affect protein functioning[17]. It has been shown that TMEM26 is an N-glycosylated protein in Jurkat T cells and breast cancer cells [7], and N-glycosylated TMEM26 can be retained on the plasma membrane for a longer time than non-glycosylated TMEM26, suggesting the important function of N-glycosylation of TMEM26 [4]. Hansel et al. [5] detected several isoproteins of TMEM26 in human breast cancer cells using western blotting, including the non-N-glycosylated protein p40TMEM26 containing 368 amino acids, and two isoproteins of p44TMEM26 and p53TMEM26 formed by N-glycosylation modification [4]. In addition, errors may exist in the predicted eight spanning transmembrane regions due to the complex protein structural configuration. For example, Town et al. [3] and Yuan et al. [18] predicted that TMEM26 may have a seven-span transmembrane structure, however, the specific transmembrane structure of TMEM26 still needs further verification. TMEM26 protein is a surface marker for beige adipocytes. The browning and differentiation of white adipose tissues into beige adipose tissues is highly effective in the treatment of obesity. TMEM26 improves glucolipid metabolism in obese patients and ameliorates cardiac metabolism in HIV-infected patients, and can be used to detect the incremental effect of browning promoters on beige cells [19-22]. The GWAS has shown that TMEM26 was associated with the depth of sleep, treatment of lung cancer with taxane, treatment of schizophrenia with antipsychotic clozapine, food allergies in European and American children, pathogenesis of diisocyanate asthma, and blood pressure elevation [23-28]. However, studies of the function of TMEM26 in the retina are rare.
There are two plexiform layers (OPL and IPL) between RGCs. Photoreceptor cells in the OPL are connected with longitudinal bipolar cells, and horizontal cells in the IPL are connected with RGCs. RGCs form synapses through interneurons in the OPL and IPL, which are connected with photoreceptors [29]. Photoreceptor cells are responsible for receiving light stimuli and transmitting light signals to nerve cells, and thus to the central nervous system via the optic nerve, resulting in the generation of visual images. In the present study, the results showed that TMEM26 was specifically expressed in the OPL and IPL of human and mouse retinas. Further studies of the function of TMEM26, especially in the retina and in vision, are of great significance.
Based on the specific expression patterns of TMEM26 in the OPL and IPL, its expression is thought to be located in the synapse, which can secrete neurotransmitters, and these neurotransmitters may affect RGCs or optic nerves. It has been reported that expression of TMEM26 was the highest in the IPL, and the IPL thickness was significantly correlated with POAG [10]; therefore, it has been speculated that TMEM26 gene expression may be associated with POAG. According to data from our group, TMEM26 gene expression is weakly correlated with POAG.
In conclusion, TMEM26 was expressed in both the OPL and IPL of the retina, and its expression was the highest in the IPL. SNP analysis confirmed that TMEM26 gene expression was weakly correlated with POAG, but its specific function remains to be determined. In the future, TMEM26-knockout mouse models should be established to study the possible function of this gene during various human diseases.
Conflicts of interest: All authors declare no conflict of interest
Author Contribution Statement: Yin Yi: topics and experimental design, experimental studies, data analyses, statistical analysis, writing, and revision; Mao Yao: experimental studies and data analyses; Yang Zhenglin: work support and experimental guidance; and Huang Lulin: topics and experimental design, experimental guidance, revision, and funding.
Acknowledgement: We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript
References
[1]Zhao KX, Liang HQ, Li QY, et al. Research progress of transmembrane protein 43[J]. J Biol, 2019, 36(4):74-77. DOI: 10.3969/j.issn.2095-1736.2019.04.074.
[2]Ryu H, Fuwad A, Yoon S, et al. Biomimetic membranes with transmembrane proteins: state-of-the-art in transmembrane protein applications[J/OL]. Int J Mol Sci, 2019, 20(6):1437[2021-09-15]. https://pubmed.ncbi.nlm.nih.gov/30901910/. DOI:10.3390/ijms2006 1437.
[3]Town L, McGlinn E, Davidson TL, et al. Tmem26 is dynamically expressed during palate and limb development but is not required for embryonic survival[J/OL]. PLoS One, 2011, 6(9):e25228[2021-09-15]. https://pubmed.ncbi.nlm.nih.gov/ 21980401/. DOI: 10.1371/journal. pone.0025228.
[4]Nass N, Dittmer A, Hellwig V, et al. Expression of transmembrane protein 26 (TMEM26) in breast cancer and its association with drug response[J/OL]. Oncotarget, 2016, 7(25):38408-38426[2021-09-17]. https://pubmed.ncbi.nlm.nih. gov/27224909/. DOI: 10.18632/ oncotarget.9493.
[5]Hansel NN, Ruczinski I, Rafaels N, et al. Genome-wide study identifies two loci associated with lung function decline in mild to moderate COPD[J]. Hum Genet, 2013, 132(1):79-90. DOI: 10.1007/s00439-012-1219-6.
[6]Wu J, Boström P, Sparks LM, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human[J]. Cell, 2012, 150(2):366-376. DOI: 10.1016/j.cell.2012.05.016.
[7]Wollscheid B, Bausch-Fluck D, Henderson C, et al. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins[J]. Nat Biotechnol, 2009, 27(4):378-386. DOI: 10.1038/nbt.1532.
[8]Zhou XM, Fan N, Liu XY. Advances in molecular genetics of glaucoma[J]. Chin J Exp Ophthalmol, 2015, 33(3):263-269. DOI: 10.3760/cma.j.issn.2095-0160. 2015.03.016.
[9]Kim KE, Park KH, Yoo BW, et al. Topographic localization of macular retinal ganglion cell loss associated with localized peripapillary retinal nerve fiber layer defect[J]. Invest Ophthalmol Vis Sci, 2014, 55(6):3501-3508. DOI: 10.1167/iovs. 14-13925.
[10]Kim EK, Park HL, Park CK. Segmented inner plexiform layer thickness as a potential biomarker to evaluate open-angle glaucoma: dendritic degeneration of retinal ganglion cell[J/OL]. PLoS One, 2017, 12(8):e0182404[2021-09-18]. https://pubmed.ncbi.nlm.nih.gov/28771565/. DOI: 10.1371/journal.pone.0182404.
[11]Chen Y, Lin Y, Vithana EN, et al. Common variants near ABCA1 and in PMM2 are associated with primary open-angle glaucoma[J]. Nat Genet, 2014, 46(10):1115-1119. DOI: 10.1038/ng.3078.
[12]Yang J, Shen HB. MemBrain-contact 2.0: a new two-stage machine learning model for the prediction enhancement of transmembrane protein residue contacts in the full chain[J]. Bioinformatics, 2018, 34(2):230-238. DOI: 10.1093/ bioinformatics/btx593.
[13]Yin X, Yang J, Xiao F, et al. MemBrain: an easy-to-use online webserver for transmembrane protein structure prediction[J/OL]. Nanomicro Lett, 2018, 10(1):2[2021-09-20]. https://pubmed.ncbi.nlm.nih.gov/30393651/. DOI: 10.1007/s40820-017-0156-2.
[14]Xiao F, Shen HB. Prediction enhancement of residue real-value relative accessible surface area in transmembrane helical proteins by solving the output preference problem of machine learning-based predictors[J]. J Chem Inf Model, 2015, 55(11):2464-2474. DOI: 10.1021/ acs.jcim.5b00246.
[15]Yang J, Jang R, Zhang Y, et al. High-accuracy prediction of transmembrane inter-helix contacts and application to GPCR 3D structure modeling[J]. Bioinformatics, 2013, 29(20):2579-2587. DOI: 10.1093/bioinformatics/btt440.
[16]Shen H, Chou JJ. MemBrain: improving the accuracy of predicting transmembrane helices[J/OL]. PLoS One, 2008, 3(6):e2399[2021-09-20]. https://pubmed.ncbi.nlm.nih.gov/ 18545655/. DOI: 10.1371/journal.pone.0002399.
[17]Krogh A, Larsson B, von Heijne G, et al. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes[J]. J Mol Biol, 2001, 305(3):567-580. DOI: 10.1006/jmbi.2000.4315.
[18]Yuan Z, Mattick JS, Teasdale RD. SVMtm: support vector machines to predict transmembrane segments[J]. J Comput Chem, 2004, 25(5):632-636. DOI: 10.1002/jcc.10411.
[19]Finlin BS, Memetimin H, Confides AL, et al. Human adipose beiging in response to cold and mirabegron[J/OL]. JCI Insight, 2018, 3(15):e121510[2021-09-21]. https://pubmed.ncbi. nlm.nih.gov/ 30089732/. DOI: 10.1172/jci.insight.121510.
[20]Lone J, Parray HA, Yun JW. Nobiletin induces brown adipocyte-like phenotype and ameliorates stress in 3T3-L1 adipocytes[J]. Biochimie, 2018, 146:97-104. DOI: 10.1016/j.biochi.2017.11.021.
[21]de Jong JM, Larsson O, Cannon B, et al. A stringent validation of mouse adipose tissue identity markers[J/OL]. Am J Physiol Endocrinol Metab, 2015, 308(12):e1085-e1105[2021-09-23]. https://pubmed.ncbi.nlm.nih.gov/25898951/. DOI: 10.1152/ajpendo.00023.2015.
[22]Torriani M, Srinivasa S, Fitch KV, et al. Dysfunctional subcutaneous fat with reduced dicer and brown adipose tissue gene expression in HIV-infected patients[J]. J Clin Endocrinol Metab, 2016, 101(3):1225-1234. DOI: 10.1210/jc.2015-3993.
[23]Spada J, Scholz M, Kirsten H, et al. Genome-wide association analysis of actigraphic sleep phenotypes in the LIFE Adult Study[J]. J Sleep Res, 2016, 25(6):690-701. DOI:10.1111/jsr.12421.
[24]Niu N, Schaid DJ, Abo RP, et al. Genetic association with overall survival of taxane-treated lung cancer patients – a genome-wide association study in human lymphoblastoid cell lines followed by a clinical association study[J/OL]. BMC Cancer, 2012, 12:422[2021-09-25]. https://pubmed.ncbi.nlm. nih.gov/23006423/. DOI: 10.1186/1471-2407-12-422.
[25]Legge SE, Hamshere ML, Ripke S, et al. Genome-wide common and rare variant analysis provides novel insights into clozapine-associated neutropenia[J]. Mol Psychiatry, 2017, 22(10):1502-1508. DOI: 10.1038/mp.2016.97.
[26]Hong X, Hao K, Ladd-Acosta C, et al. Genome-wide association study identifies peanut allergy-specific loci and evidence of epigenetic mediation in US children[J/OL]. Nat Commun, 2015, 6:6304[2021-09-25]. https://pubmed.ncbi. nlm.nih.gov/25710614/. DOI: 10.1038/ncomms7304.
[27]Yucesoy B, Kaufman KM, Lummus ZL, et al. Genome-wide association study identifies novel loci associated with diisocyanate-induced occupational asthma[J]. Toxicol Sci, 2015, 146(1):192-201. DOI: 10.1093/toxsci/kfv084.
[28]Newton-Cheh C, Johnson T, Gateva V, et al. Genome-wide association study identifies eight loci associated with blood pressure[J]. Nat Genet, 2009, 41(6):666-676. DOI: 10.1038/ng.361.
[29]Kim EK, Park HL, Park CK. Relationship between retinal inner nuclear layer thickness and severity of visual field loss in glaucoma[J/OL]. Sci Rep, 2017, 7(1):5543[2021-09-26]. https://pubmed.ncbi.nlm.nih.gov/28717139/. DOI: 10.1038/s41598-017-05282-4.