·Experimental Research·
Differential expression analysis of circular RNA in human retinal vascular endothelial cells induced by high glucose
Jia Yangxue1, Wang Zhiling2, Wei Yingying2, Zhu Kai2, Gu Yonghao2
1Department of Ophthalmology, Second Affiliated Hospital of Wannan Medical College, Wuhu 241000, China; 2Department of Ophthalmology, Provincial Hospital Affiliated to Anhui Medical University, Hefei 230001, China
Corresponding author: Gu Yonghao, Email: aerolplane@hotmail.com
[Abstract] [View PDF in English] [View PDF in Chinese] [Read Full Text]
Objective To investigate the differential expression profile of circular RNA (circRNA) in high glucose-cultured human retinal vascular endothelial cells (HRVECs).
Methods The HRVECs were divided into high glucose group, normal glucose group and hypertonic control group, and were cultures in 25 mmmol/L glucose medium, 5.5 mmol/l glucose mediumn and 19.5 mmol/L mannitol+5.5 mmol/l glucose medium for 24 hours accordingly. The differentially expressed circRNA molecules between high glucose group and normal glucose group were screened by circRNA microarray analysis. The expression of the most significant differentially expressed circRNAs in different groups was verified by real-time quantitative PCR. The possible microRNA (miRNA) targets were analyzed.
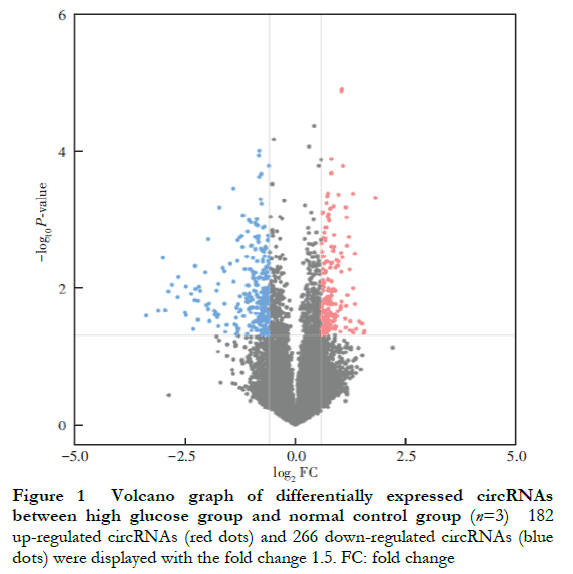
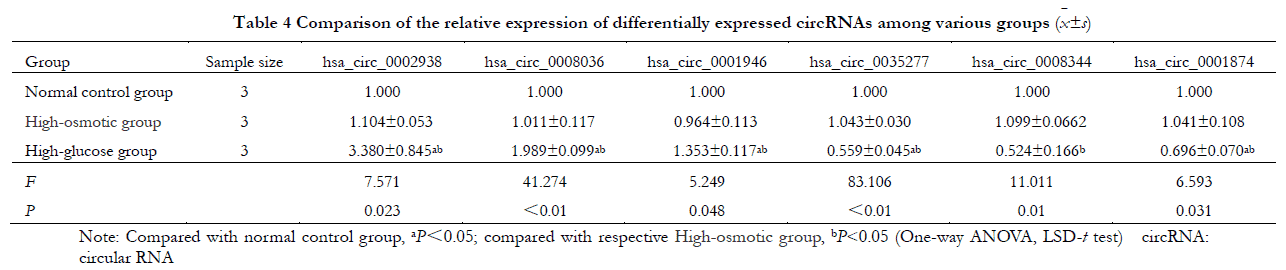
Results It is found that 448 circRNAs were differentially expressed (FC≥1.5 or FC≤0.67, P<0.05) in high glucose-cultured HRVECs, among which 182 were up-regulated and 266 were down-regulated. The top 3 significantly up-regulated circRNAs were hsa_circ_0002938, hsa_circ_0008036, and hsa_circ_0001946, and the top 3 significantly down-regulated circRNAs were hsa_circ_0035277, hsa_circ_0008344, and hsa_circ_0001874. Compared with normal control group and hypertonic control group, the relative expressions of top 3 up-regulated circRNAs were significantly enhanced and the relative expressions of top 3 down-regulated circRNAs were significantly reduced in high glucose group, showing statistically significant differences (all at P<0.05). No significant difference was found in the differentially expressed circRNAs between normal control group and hypertonic control group (all at P>0.05).
Conclusions CircRNAs are differentially expressed in high glucose-cultured HRVECs, and the differentially expressed circRNAs may be involved in the regulatory mechanism of diabetic retinopathy.
Key words Diabetic retinopathy; Circular RNA; Microarray; Target gene
Fund program: Anhui Natural Science Foundation (1908085MH254)
DOI: 10.3760/cma.j.cn115989-20200602-00390
Diabetic retinopathy (DR) is a major sight-threatening microvascular complication of diabetes. The overall prevalence of DR among diabetic patients is about 23% in China. Studies show that the prevalence of DR in patients who have been diagnosed with type 1 and type 2 diabetes for at least 15 years is 98% and 78%, respectively. Given a lack of effective means to prevent and treat DR, it is important to investigate the pathogenesis of DR to enable the development of novel therapeutic interventions. Vascular endothelial cell dysfunction and metabolic changes in the retinal microenvironment have been implicated in DR pathogenesis. Therefore, prevention of these changes can potentially reduce the risk of diabetic vascular complications. Circulac RNA (circRNA) represents a class of non-coding RNA that is widely found in eukaryotic organisms, which is characterized by high stability, conservativeness, certain time sequence and tissue specificity. The major functions of circRNAs include (1) competing with microRNAs (miRNAs) for binding; (2) interacting with proteins through RNA-binding proteins; (3) regulating the expression of parental genes by modulating the transcription of RNA polymerase; (4) mediating the formation of pseudogenes through selective splicing or base complementary pairing; (5) and acting as mRNA-encoded proteins. Growing evidence increasingly implicates circRNAs in the pathology of lung, esophageal, gastric, colorectal, pancreatic and hepatocellular carcinomas. As such, circRNAs is a potential target for the early diagnosis and treatment of these cancers. Moreover, it has also been suggested that circRNA influences the secretory function of pancreatic islet cells by regulating the expression of downstream target genes, and thus playing a role in the development of diabetes and its complications. A comparative study conducted by our group using circRNA microarrays of plasma from five patients with proliferative DR, five patients with type 2 diabetes mellitus without significant fundus lesions, and five age-matched normal subjects revealed significant changes in plasma circRNAs profile in patients with proliferative DR — confirming the potential role of circRNAs in the pathogenesis of DR. Analysis of plasma samples is known to be inaccurate owing to the vulnerability of its components to the overall body conditions. In contrast, direct cellular assays are more reliable in detecting the effect of high glucose stimulation on circRNA expression in retinal vascular endothelial cells. In this study, we aim to identify the differentially expressed circRNAs using circRNA microarray between normal and high glucose cultured human retinal vascular endothelial cells (HRVECs) before validating them by real-time fluorescence quantitative PCR. We also aim to investigate their possible target miRNA, with a view to ultimately enabling more effective diagnosis and treatment of DR.
1 Materials and methods
1.1 Materials
1.1.1 Cell source HRVECc was obtained from Shenzhen Haodi Huatuo Biotechnology Co.
1.1.2 Reagents and instruments DMEM low sugar medium (HyC/ne, USA), Trypsin, fetal bovine serum (FBS), Penicillin-streptomycin mixture (Gibco, USA), Mannitol reagent (Shanghai Zhangyun Chemical Co., Ltd.), Trizoi reagent (AmbRn, USA), Reverse transcription kit (ABclonai, USA), circRNA GeneChip (Arraystar, USA), SuperRed Gel/ed Nucleic Acid Dye (BRsearch Technologies, USA), Fluorescent quantitative PCR reagent Geniouc 2X SYBR Green Fast qPCR (ABclonai, USA), Low-speed centrifuge (Anhui Zhongke Zhongjia Scientific Instruments Co., Ltd.), CO2 cell culture chamber, real-time fluorescence quantitative PCR instrument (Thermo, USA), DYY-6C electrophoresis instrument (Beijing Liuyi Biotechnology Co., Ltd.), Nanodrop micro-optical spectrophotometer (Thermo, USA), UV spectrophotometer (Shanghai Sile Instruments Co., Ltd.) and Gel imager (Anhui Zhongke Zhongjia Scientific Instruments Co., Ltd.).
1.2 Methods
1.2.1 Culture and Grouping of HRVECs HRVECs were cultured and passaged in DMEM low sugar medium containing 10% FBS by volume and 100 U/ml of penicillin/streptomycin mixture at 37℃and 5% CO2 by volume in a constant temperature CO2 incubator, with fluid changes every 2~3 days. After growing to 80% confluence, the cells were passaged and cultured in groups, namely (1) normal control cells in 5.5 mmol/L glucose medium; (2) hypertonic control cells in 19.5 mmol/L mannitol and 5.5 mmol/L glucose medium; (3) and high glucose cells in 25 mmo/L glucose medium. Each group of cells was incubated in a cell thermostat at 37℃ for 24 hours. The experiments were repeated three times independently.
1.2.2 Detection of circRNA expression in cells by circRNA microarray and screening of differential genes The cells of normal control and high glucose groups were washed three times with pre-cooled phosphate buffer, before being lysed by adding Tczol to extract the total RNA from the samples. The purity and concentration of nucleic acids were determined by UV spectrophotometer, i.e. the value of A260/A280 should be 1-8 – 2.0, and the total amount of RNA should reach 500 ug. The purity and integrity of RNA were determined by denaturing agarose gel electrophoresis with formaldehyde electrophoresis reagent. The assays were performed by Shanghai Kangcheng Bioengineering Co., Ltd. using circRNA microarray V2.0. The total RNA extracted from the normal control and high glucose groups was treated with RNase R. The cDNA was synthesized by reverse transcription and labelled with fluorescence using the Array star Supec RNA Labeling kit. The fluorescent labeling efficiency was determined by Nanodrop, and the labeled probes were hybridized with the high-density genomic microarray under standard conditions. The GenePia 4000B microarray scanner was used to detect the fluorescence intensity of the microarrays. GenePix Pro6 software was used to convert the results into digital data for storage purposes. The raw data were normalized using the R limma package, and the differentially expressed circRNAs in the two groups of samples were identified according to the fold change (FC)≥1.5 or FC≤0.67 and P<0.05.
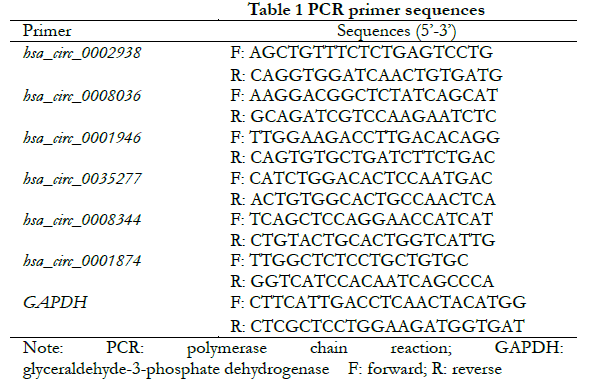
1.2.3 Validation of differentially expressed circRNA using real-time fluorescent quantitative PCR The top 3 circRNAs ranked for FC were selected from the up- and down-regulated differentially expressed cicRNAs, respectively. Firstly, the full-length sequences of circRNAs were obtained from the circRNA database (http://www.circbase.org/). The primers were designed upstream and downstream across the junction site of circRNA according to the divergent primer design principle to ensure that the amplified products contained the junction site. The specific primer sequences of each target circRNA are shown in Table 1. The primers were synthesized by Hefei General Biosystems with GAPDH as the internal reference. Cells from normal control, hyperosmotic control and high glucose groups were collected and treated with Trizol to extract total RNA. cDNA was obtained by quantitative reverse transcription, and primers were amplified by real-time fluorescence quantitative PCR. A total of 50 μl of reaction system was used, including 1 μl of forward reverse primer alongside 1 μl of reverse primer, 2 μl of cDNA, 21 μl of RNase-free water, and 25 μl of qRT-PCR enzyme. 15 μl of the mixture was added to three wells of a 96-well plate — centrifuged — before running the program in a qPCR instrument. The reaction conditions for real-time fluorescence PCR were as follows: (1) pre-denaturation at 95 ℃ for 3 min; (2) denaturation at 95 ℃ for 5 s; (3) annealing at 60 ℃; (4) and extension for 34 s. A total of 40 cycles were performed. The melting curves (95 ℃ 1 s, 60 ℃ 20 s, 95 ℃ 1 s) were used to analyze the amplified product. The relative expression of each target gene was calculated using the 2-∆∆Ct method.
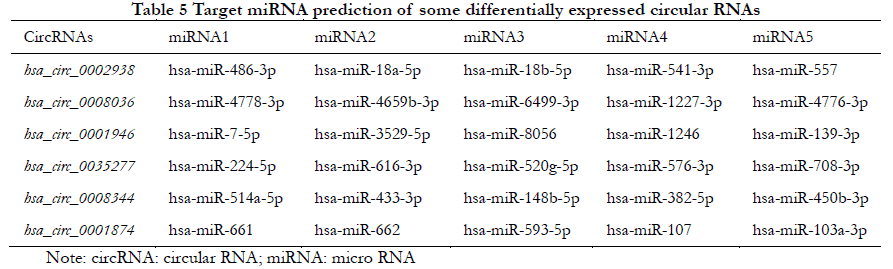
1.2.4 Prediction of miRNA binding sites The Circinteractome database (https://circinteractome.nR.nih.gov/index.html) was used to predict the potential downstream miRNA targets of the six most significantly differentially expressed circRNA molecules based on their tightness of binding.
1.3 Statistical Analysis
SPSS 19.0 software was used for statistical analysis. The data of measurement data were confirmed to be normally distributed using Shapiro-Wilk test and expressed as `x±s. The overall difference of circRNA relative expression among the three groups was compared with one-way ANOVA, while the LSD-t test was used in a two-way comparison between groups. P<0.05 was considered to be statistically significance.
2 Results
2.1 Screening of differentially expressed circRNAs
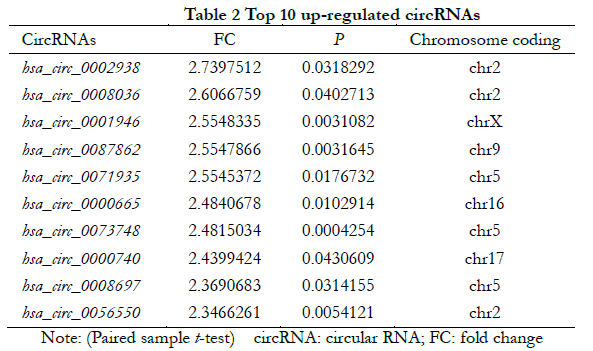
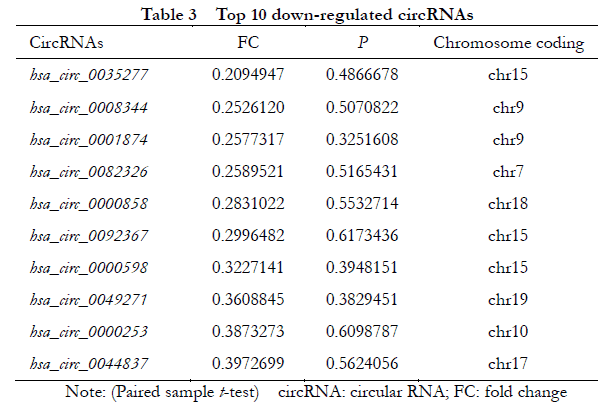
A total of 448 differentially expressed circRNAs were identified, of which 182 were upregulated and 266 were downregulated (Figure 1 ). The 10 most significantly upregulated circRNAs were hsa_circ_0002938, hsa_circ_0008036, hsa_circ_0001946, hsa_circ_0087862, hsa_circ_0071935, hsa_circ_0000665, hsa_circ_0073748, hsa_circ_0000740, hsa_circ_0008697, and hsa_circ_0056550 (Table 2). The 10 most significantly downregulated circRNAs were hsa_circ_0035277, hsa_circ_0008344, hsa_circ_0001874, hsa_circ_0082326, hsa_circ_0000858, hsa_circ_0092367, hsa_circ_0000598, hsa_circ_0049271, hsa_circ_0000253, and hsa_circ_0044837 (Table 3).
2.2 Validation of differential circRNA using real-time fluorescence quantitative PCR
Overall, the relative expressions of the three significantly up-regulated circRNAs (hsa_circ_0002938, hsa_circ_0008036 and hsa_circ_0001946) as well as the three significantly down-regulated circRNAs (hsa_circ_0035277, hsa_circ_0008344 and hsa_circ_0001874) in the normal control, hyperosmolar control and hyperglycemic groups differed statistically significantly (F=7.571, 41.274, 5.249, 83.106, 11.011, 6.593, all P<0.05). However, the differences in the relative expression of circRNAs between normal control and hyperosmolar control groups were not statistically significant (all P>0.05). Compared with the normal control group and the hyperosmolar control group, the relative expressions of hsa_circ_0002938, hsa_circ_0008036 and hsa_circ_0001946 in the high glucose group were significantly higher, while the relative expressions of hsa_circ_0035277, hsa_circ_0008344 and hsa_circ_0001874 were significantly lower (all P<0. 05) (Table 4).
2.3 Target gene prediction of partially differentially expressed circRNA
The five most likely target miRNAs for each differentially expressed circRNA are listed in Table 5 according to the number of binding sites and the degree of tightness.
3 Discussion
Zhang et al. identified 529 differentially expressed circRNAs in the retinal tissues of three normal subjects and two diabetic patients. They suggested that has_circRNA_0005015 might bind to miR-519d-3p and regulate vascular endothelial cell function. CircRNA crcHIPK3 inhibited miRNA-30a-3p expression, resulting in increased expressions of vascular endothelial growth factor C, Wnt2 and frizzled class receptor 4 (FZD4). This in turn caused endothelial cell proliferation and vascular dysfunction. Under hypoxic and high-glucose conditions, the expression of circRNA-ZNF609 was significantly upregulated. It binds to the endogenous miR-615-5p, thereby inhibiting miR-615-5p activity and enhancing HRVECs viability, mobility and lumen formation. Furthermore, high-throughput sequencing of differentially expressed circRNAs in the vitreous specimens from DR patients and non-diabetic patients in addition to high-glucose cultured human umbilical vein vascular endothelial cells confirmed that circRNAs regulate the gene expression associated with vascular endothelial function and angiogenesis by targeting miRNAs. Liu et al. studied circRNA-cPWWP2A in pericytes and found that it bound to miR579 and improved retinal leakage in an animal model of diabetes. In contrast, we did not find any of the same differentially expressed circRNA molecules as in these studies. This may be attributable to the difference in specimens used.
In this study, 448 differentially expressed circRNAs were identified by comparing the circRNA expression profiles of high glucose and normal culture HRVECs. Six significantly differentially expressed circRNAs were validated using real-time fluorescence quantitative PCR, and the results were consistent with the microarray assay, indicating that microarray assay is reliable. Among them, the results pertaining to hsa_circ_0001946 were consistent with our group’s previous study on differentially expressed circRNA in the plasma of patients with proliferative DR.
The predicted target miRNAs with strong binding to hsa_circ_0001946 included hsa-miR-7-5p, hsa-miR-3529-5p, hsa-miR- 8056, hsa-miR-1246, and hsa-miR-139-3p. Hsa-miR-7-5p binds to long-stranded non-coding small nucleolar RNA host gene 16 (ncRNA small nucleolar RNA host gene 16, SNHG16) in the high-glucose state, making SNHG16 an endogenous RNA competing for interleukin-1 receptor-related kinase 1 and insulin receptor substrate 1, thereby inducing retinal microvascular endothelial cell dysfunction and inhibiting endothelial cell proliferation. The expression of hsa-miR-1246 was significantly upregulated in endothelium-derived exosomes, which promotes fibroblast to endothelial cell phenotype transformation and angiogenesis. In addition, hsa-miR-1246 is involved in lipopolysaccharide-induced inflammatory response and apoptosis in endothelial cells, whereas hsa-miR-139-3p is involved in the development and progression of cervical cancer through the activation of oncoproteins p53, p21 and p16.
MiR-486-3p is one of the predicted target miRNAs of hsa_circ_0002938. Its upregulation has been reported to suppress oxidative stress, inflammation and apoptosis in DR mice and promote muller cell proliferation under high glucose status. MiR-224-5p is one of the predicted target miRNAs of hsa_circ_ 0035277, and it has been implicated in the inflammatory response in the hippocampus of type 2 diabetic patients through the NLRP3 pathway. These downstream miRNAs are involved in oxidative stress, inflammation and other processes in the pathogenesis of DR, which affect endothelial cell proliferation, migration and lumen formation capacity as well as the expression of vascular endothelial growth factor. The activation and invasion of endothelial cells is one of the key processes in the neovascularization of DR. Therefore, we hypothesize that differentially expressed circRNAs such as hsa_circ_0001946 may lead to retinal endothelial cell dysfunction by binding to the corresponding target miRNAs, and thus influencing the onset and progression of DR. That said, further studies are needed to confirm our results.
In summary, the present study obtained the differential expression profiles of circRNAs in high glucose cultured HRVECs. After validating six of the most significantly differentially expressed circRNAs (hsa_circ_0002938, hsa_circ_0008036, hsa_circ_0001946, hsa_circ_0035277, hsa_circ_0008344 and hsa_circ_0001874), their possible target miRNAs were predicted. Our results indicate that these circRNAs may influence the onset and development of DR via miRNAs. The present study also provides new evidence for the involvement of circRNAs in DR, but the roles of these circRNA molecules in DR courses and the underlying mechanisms remain elusive.
Conflicts of interest None declared.
Acknowledgements The assistance provided by School of Life Sciences, University of Science and Technology of China and Aksomics Inc. are greatly appreciated.
Author contributions Jia Yangxue collected the data, performed the analysis and wrote the paper. Wamg Zhiling, Wei Yingying and Zhu Kai collected and analyzed the data. Gu Yonghao designed the analysis and revised the paper.
References
[1] Wang F, Wang CF, Yan JL. Survey of diabetic retinopathy of the diabetic population over 45 years old[J]. Chin J Exp Ophthalmol, 2013, 31(8):783-787. DOI: 10.3760/cma.j.issn.2095-0160.2013.08.018.
[2] Roy MS, Klein R, O’Colmain BJ, et al. The prevalence of diabetic retinopathy among adult type 1 diabetic persons in the United States[J]. Arch Ophthalmol, 2004, 122(4):546-551. DOI: 10.1001/archopht.122.4.546.
[3] Liao Y, Chen H. Effects of complement dysfunction on diabetic retinopathy[J]. Chin J Exp Ophthalmol, 2020, 38(1): 68-72. DOI: 10.3760/cma.j.issn.2095-0160.2020.01.014.
[4] Sorrentino FS, Matteini S, Bonifazzi C, et al. Diabetic retinopathy and endothelin system: microangiopathy versus endothelial dysfunction[J]. Eye (Lond), 2018, 32(7):1157-1163. DOI: 10.1038/s41433-018-0032-4.
[5] Li JY, Huang WQ, Tu RH, et al. Resveratrol rescues hyperglycemia-induced endothelial dysfunction via activation of Akt[J]. Acta Pharmacol Sin, 2017, 38(2):182-191. DOI: 10.1038/aps.2016.109.
[6] Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges[J]. Nature, 2013, 495(7441):384-388. DOI: 10.1038/nature11993.
[7] Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus[J]. Nat Struct Mol Biol, 2015, 22(3):256-264. DOI: 10.1038/nsmb.2959.
[8] Dong R, Zhang XO, Zhang Y, et al. CircRNA-derived pseudogenes[J]. Cell Res, 2016, 26(6):747-750. DOI: 10.1038/cr.2016.42.
[9] Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition[J]. Nature, 2014, 505(7483):344-352. DOI: 10.1038/nature12986.
[10] Cao X, Zhang X, Jiang PC. Research progress of circular RNA in malignant tumors[J]. Cancer Prog, 2020, 18(13):1297-1300,1400. DOI: 10.11877/j.issn.1672-1535.2020.18.13.01.
[11] Bai Y, Liu WH. Correlation of circular RNAs with diabetes mellitus and its complications[J]. J Clin Nephrol, 2020, 20(5):421-424. DOI: 10.3969/j.issn.1671-2390.2020.05.014.
[12] Gu Y, Ke G, Wang L, et al. Altered expression profile of circular RNAs in the serum of patients with diabetic retinopathy revealed by microarray[J]. Ophthalmic Res, 2017, 58(3):176-184. DOI: 10.1159/000479156.
[13] Zhang SJ, Chen X, Li CP, et al. Identification and characterization of circular RNAs as a new class of putative biomarkers in diabetes retinopathy[J]. Invest Ophthalmol Vis Sci, 2017, 58(14):6500-6509. DOI: 10.1167/iovs.17-22698.
[14] Shan K, Liu C, Liu BH, et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus[J]. Circulation, 2017, 136(17):1629-1642. DOI: 10.1161/CIRCULATIONAHA.117.029004.
[15] Liu C, Yao MD, Li CP, et al. Silencing of circular RNA-ZNF609 ameliorates vascular endothelial dysfunction[J]. Theranostics, 2017, 7(11):2863-2877. DOI: 10.7150/thno.19353.
[16] He M, Wang W, Yu H, et al. Comparison of expression profiling of circular RNAs in vitreous humour between diabetic retinopathy and non-diabetes mellitus patients[J]. Acta Diabetol, 2020, 57(4):479-489. DOI: 10.1007/s00592-019-01448-w.
[17] Jin G, Wang Q, Hu X, et al. Profiling and functional analysis of differentially expressed circular RNAs in high glucose-induced human umbilical vein endothelial cells[J]. FEBS Open Bio, 2019, 9(9):1640-1651. DOI: 10.1002/2211-5463.12709.
[18] Liu C, Ge HM, Liu BH, et al. Targeting pericyte-endothelial cell crosstalk by circular RNA-cPWWP2A inhibition aggravates diabetes-induced microvascular dysfunction[J]. Proc Natl Acad Sci U S A, 2019, 116(15):7455-7464. DOI: 10.1073/pnas.1814874116.
[19] Cai F, Jiang H, Li Y, et al. Upregulation of long non-coding RNA SNHG16 promotes diabetes-related RMEC dysfunction via activating NF-κB and PI3K/AKT pathways[J]. Mol Ther Nucleic Acids, 2021, 24:512-527. DOI: 10.1016/j.omtn.2021.01.035.
[20] Huang Y, Chen L, Feng Z, et al. EPC-derived exosomal miR-1246 and miR-1290 regulate phenotypic changes of fibroblasts to endothelial cells to exert protective effects on myocardial infarction by targeting ELF5 and SP1[J/OL]. Front Cell Dev Biol, 2021, 9:647763[2022-03-11]. http://www.ncbi.nlm.nih.gov/pubmed/34055778. DOI: 10.3389/fcell.2021.647763.
[21] Fang Y, Gao F, Hao J, et al. microRNA-1246 mediates lipopolysaccharide-induced pulmonary endothelial cell apoptosis and acute lung injury by targeting angiotensin-converting enzyme 2[J]. Am J Transl Res, 2017, 9(3):1287-1296.
[22] Sannigrahi MK, Sharma R, Singh V, et al. Role of host miRNA Hsa-miR-139-3p in HPV-16-induced carcinomas[J]. Clin Cancer Res, 2017, 23(14):3884-3895. DOI: 10.1158/1078-0432.CCR-16-2936.
[23] Li W, Jin L, Cui Y, et al. Bone marrow mesenchymal stem cells-induced exosomal microRNA-486-3p protects against diabetic retinopathy through TLR4/NF-κB axis repression[J]. J Endocrinol Invest, 2021, 44(6):1193-1207. DOI: 10.1007/s40618-020-01405-3.
[24] Du P, Wang J, Han Y, et al. Blocking the LncRNA MALAT1/miR-224-5p/NLRP3 axis inhibits the hippocampal inflammatory response in T2DM with OSA[J/OL]. Front Cell Neurosci, 2020, 14:97[2022-0-13]. http://www.ncbi.nlm.nih.gov/pubmed/32477065. DOI: 10.3389/fncel.2020.00097.